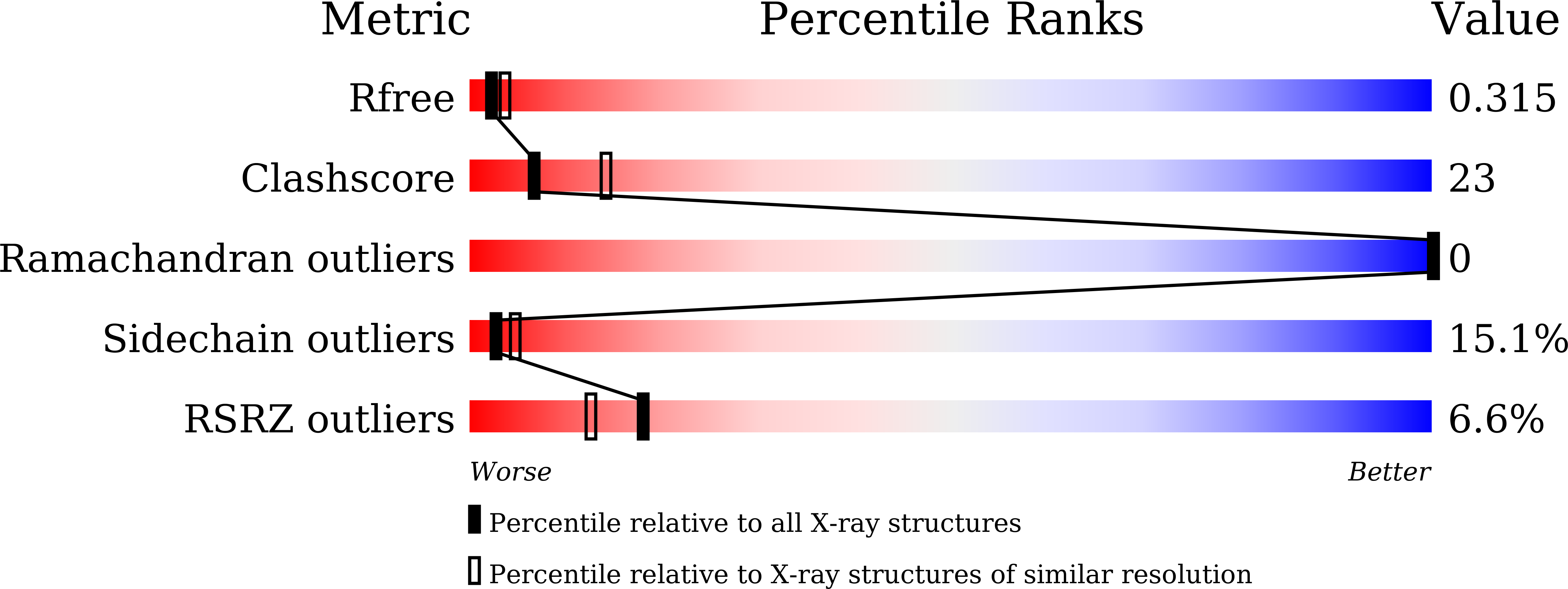
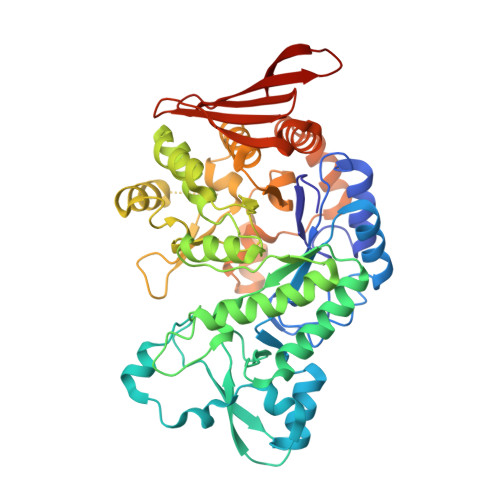
Protein Engineering of Glucosylglycerol Phosphorylase Facilitating Efficient and Highly Regio- and Stereoselective Glycosylation of Polyols in a Synthetic System.
Zhang, T., Liu, P., Wei, H., Sun, X., Zeng, Y., Zhang, X., Cai, Y., Cui, M., Ma, H., Liu, W., Sun, Y., Yang, J.(2022) ACS Catal : 15715-15727