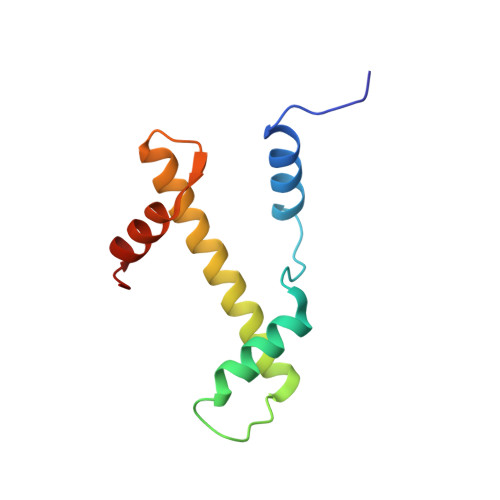
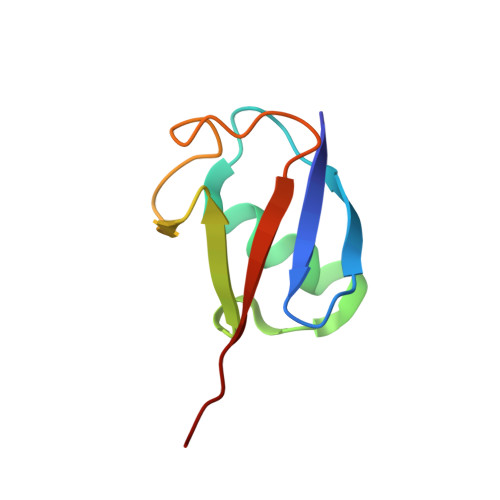
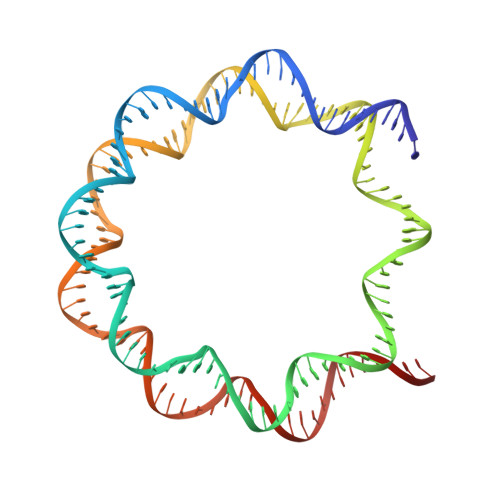
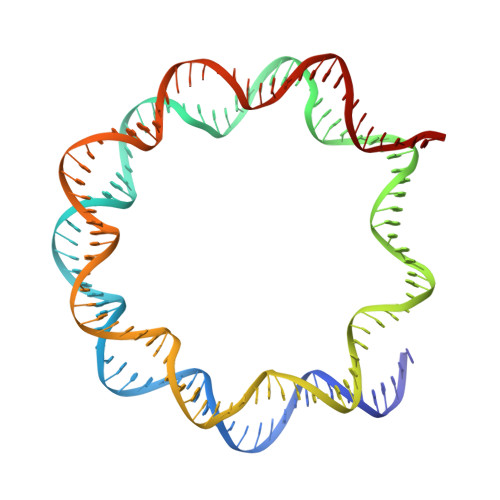
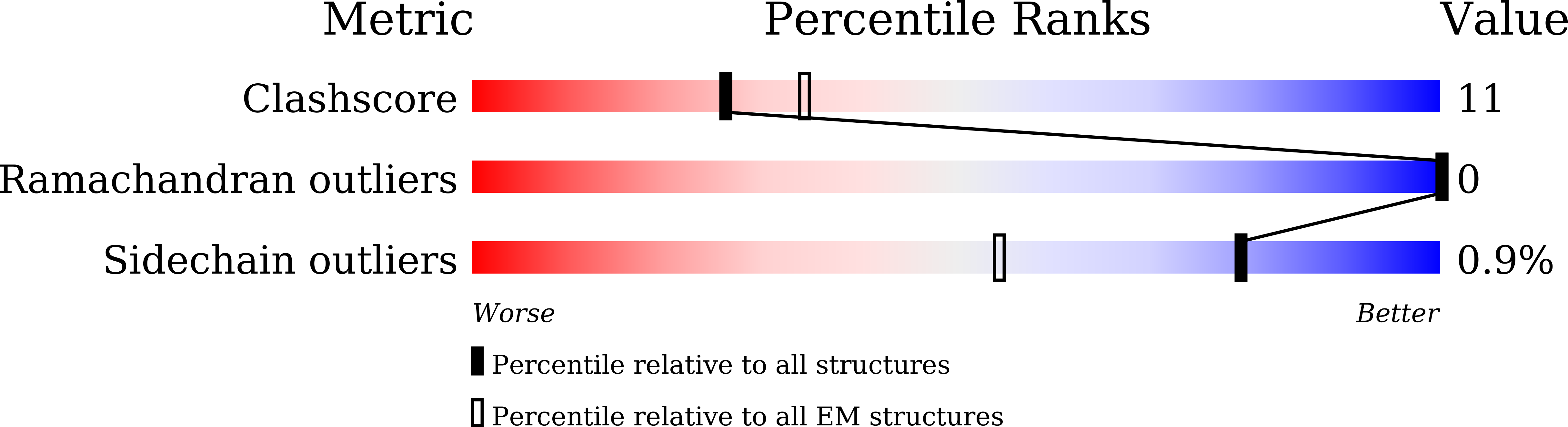H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Ai, H., Sun, M., Liu, A., Sun, Z., Liu, T., Cao, L., Liang, L., Qu, Q., Li, Z., Deng, Z., Tong, Z., Chu, G., Tian, X., Deng, H., Zhao, S., Li, J.B., Lou, Z., Liu, L.(2022) Nat Chem Biol 18: 972-980
- PubMed: 35739357
- DOI: https://doi.org/10.1038/s41589-022-01067-7
- Primary Citation of Related Structures:
7XCR, 7XCT, 7XD0, 7XD1 - PubMed Abstract:
Ubiquitination-dependent histone crosstalk plays critical roles in chromatin-associated processes and is highly associated with human diseases. Mechanism studies of the crosstalk have been of the central focus. Here our study on the crosstalk between H2BK34ub and Dot1L-catalyzed H3K79me suggests a novel mechanism of ubiquitination-induced nucleosome distortion to stimulate the activity of an enzyme. We determined the cryo-electron microscopy structures of Dot1L-H2BK34ub nucleosome complex and the H2BK34ub nucleosome alone. The structures reveal that H2BK34ub induces an almost identical orientation and binding pattern of Dot1L on nucleosome as H2BK120ub, which positions Dot1L for the productive conformation through direct ubiquitin-enzyme contacts. However, H2BK34-anchored ubiquitin does not directly interact with Dot1L as occurs in the case of H2BK120ub, but rather induces DNA and histone distortion around the modified site. Our findings establish the structural framework for understanding the H2BK34ub-H3K79me trans-crosstalk and highlight the diversity of mechanisms for histone ubiquitination to activate chromatin-modifying enzymes.
Organizational Affiliation:
Department of Chemistry, Tsinghua-Peking Joint Center for Life Sciences, MOE Key Laboratory of Bioorganic Phosphorus Chemistry and Chemical Biology, Tsinghua University, Beijing, China.