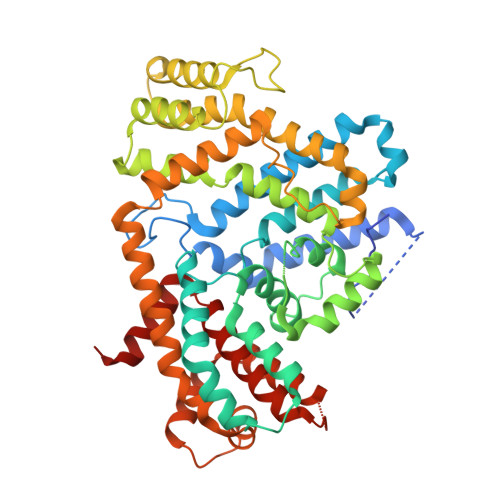
Structural analysis of the dNTP triphosphohydrolase PA1124 from Pseudomonas aeruginosa.
Oh, H.B., Lee, K.C., Park, S.C., Song, W.S., Yoon, S.I.(2022) Biochem Biophys Res Commun 589: 78-84
- PubMed: 34894560
- DOI: https://doi.org/10.1016/j.bbrc.2021.12.002
- Primary Citation of Related Structures:
7W1F - PubMed Abstract:
dNTP triphosphohydrolase (TPH) belongs to the histidine/aspartate (HD) superfamily and catalyzes the hydrolysis of dNTPs into 2'-deoxyribonucleoside and inorganic triphosphate. TPHs are required for cellular dNTP homeostasis and DNA replication fidelity and are employed as a host defense mechanism. PA1124 from the pathogenic Pseudomonas aeruginosa bacterium functions as a dGTP and dTTP triphosphohydrolase. To reveal how PA1124 drives dNTP hydrolysis and is regulated, we performed a structural study of PA1124. PA1124 assembles into a hexameric architecture as a trimer of dimers. Each monomer has an interdomain dent where a metal ion is coordinated by conserved histidine and aspartate residues. A structure-based comparative analysis suggests that PA1124 accommodates the dNTP substrate into the interdomain dent near the metal ion. Interestingly, PA1124 interacts with ssDNA, presumably as an allosteric regulator, using a positively charged intersubunit cleft that is generated via dimerization. Furthermore, our phylogenetic analysis highlights similar or distinct oligomerization profiles across the TPH family.
Organizational Affiliation:
Division of Biomedical Convergence, College of Biomedical Science, Kangwon National University, Chuncheon, 24341, South Korea.