Highly efficient production of rebaudioside D enabled by structure-guided engineering of bacterial glycosyltransferase YojK.
Guo, B., Hou, X., Zhang, Y., Deng, Z., Ping, Q., Fu, K., Yuan, Z., Rao, Y.(2022) Front Bioeng Biotechnol 10: 985826-985826
- PubMed: 36091437
- DOI: https://doi.org/10.3389/fbioe.2022.985826
- Primary Citation of Related Structures:
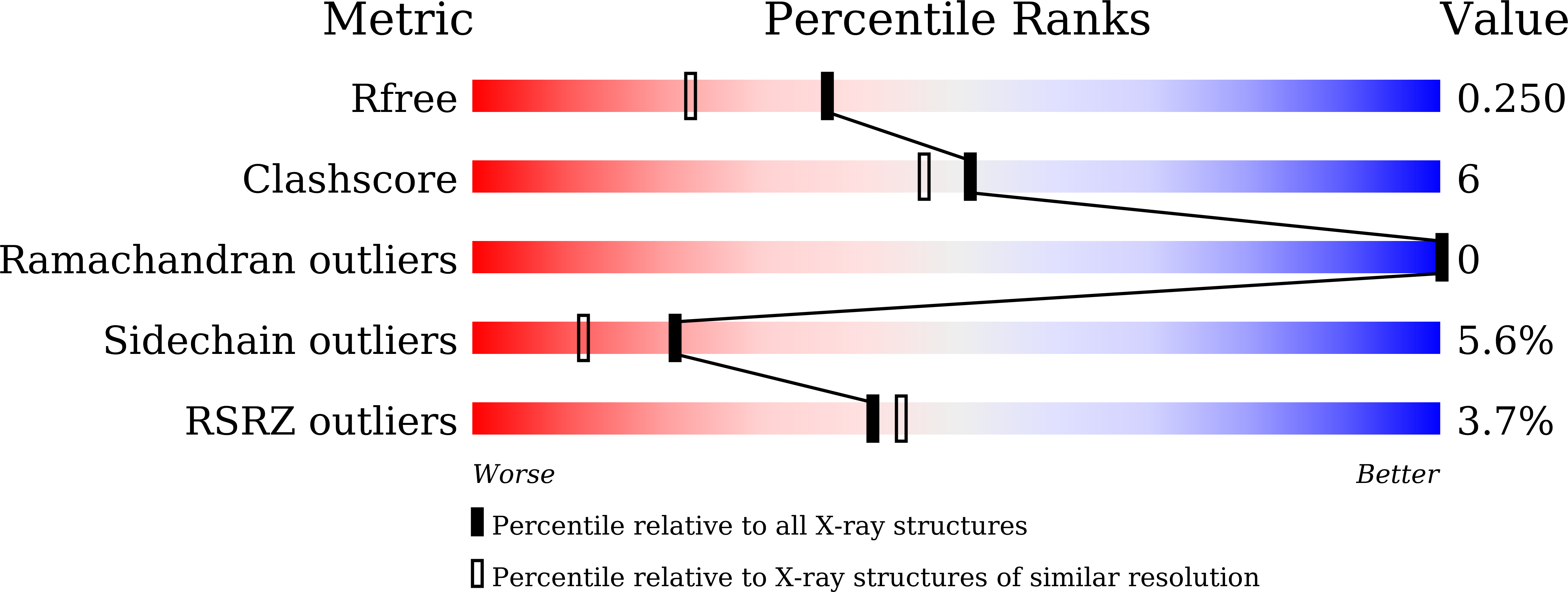
7VM0 - PubMed Abstract:
Owing to zero-calorie, high-intensity sweetness and good taste profile, the plant-derived sweetener rebaudioside D (Reb D) has attracted great interest to replace sugars. However, low content of Reb D in stevia rebaudiana Bertoni as well as low soluble expression and enzymatic activity of plant-derived glycosyltransferase in Reb D preparation restrict its commercial usage. To address these problems, a novel glycosyltransferase YojK from Bacillus subtilis 168 with the ability to glycosylate Reb A to produce Reb D was identified. Then, structure-guided engineering was performed after solving its crystal structure. A variant YojK-I241T/G327N with 7.35-fold increase of the catalytic activity was obtained, which allowed to produce Reb D on a scale preparation with a great yield of 91.29%. Moreover, based on the results from molecular docking and molecular dynamics simulations, the improvement of enzymatic activity of YojK-I241T/G327N was ascribed to the formation of new hydrogen bonds between the enzyme and substrate or uridine diphosphate glucose. Therefore, this study provides an engineered bacterial glycosyltransferase YojK-I241T/G327N with high solubility and catalytic efficiency for potential industrial scale-production of Reb D.
Organizational Affiliation:
Key Laboratory of Carbohydrate Chemistry and Biotechnology, Ministry of Education, School of Biotechnology, Jiangnan University, Wuxi, China.