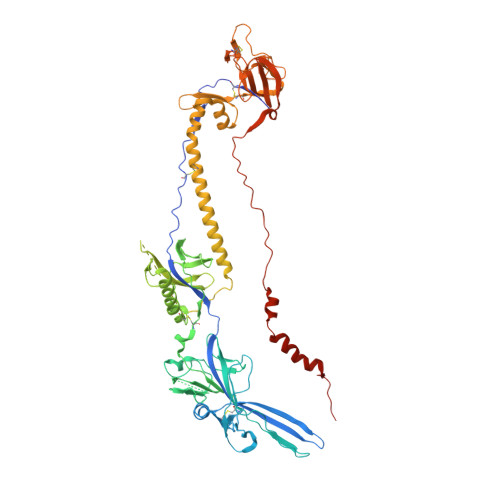
A non-neutralizing glycoprotein B monoclonal antibody protects against herpes simplex virus disease in mice.
Kuraoka, M., Aschner, C.B., Windsor, I.W., Mahant, A.M., Garforth, S.J., Kong, S.L., Achkar, J.M., Almo, S.C., Kelsoe, G., Herold, B.C.(2023) J Clin Invest 133
- PubMed: 36454639
- DOI: https://doi.org/10.1172/JCI161968
- Primary Citation of Related Structures:
7UHZ, 7UI0 - PubMed Abstract:
There is an unmet need for monoclonal antibodies (mAbs) for prevention or as adjunctive treatment of herpes simplex virus (HSV) disease. Most vaccine and mAb efforts focus on neutralizing antibodies, but for HSV this strategy has proven ineffective. Preclinical studies with a candidate HSV vaccine strain, ΔgD-2, demonstrated that non-neutralizing antibodies that activate Fcγ receptors (FcγRs) to mediate antibody-dependent cellular cytotoxicity (ADCC) provide active and passive protection against HSV-1 and HSV-2. We hypothesized that this vaccine provides a tool to identify and characterize protective mAbs. We isolated HSV-specific mAbs from germinal center and memory B cells and bone marrow plasmacytes of ΔgD-2-vaccinated mice and evaluated these mAbs for binding, neutralizing, and FcγR-activating activity and for protective efficacy in mice. The most potent protective mAb, BMPC-23, was not neutralizing but activated murine FcγRIV, a biomarker of ADCC. The cryo-electron microscopic structure of the Fab-glycoprotein B (gB) assembly identified domain IV of gB as the epitope. A single dose of BMPC-23 administered 24 hours before or after viral challenge provided significant protection when configured as mouse IgG2c and protected mice expressing human FcγRIII when engineered as a human IgG1. These results highlight the importance of FcR-activating antibodies in protecting against HSV.
Organizational Affiliation:
Department of Immunology, Duke University School of Medicine, Durham, North Carolina, USA.