Evasion of cGAS and TRIM5 defines pandemic HIV.
Zuliani-Alvarez, L., Govasli, M.L., Rasaiyaah, J., Monit, C., Perry, S.O., Sumner, R.P., McAlpine-Scott, S., Dickson, C., Rifat Faysal, K.M., Hilditch, L., Miles, R.J., Bibollet-Ruche, F., Hahn, B.H., Boecking, T., Pinotsis, N., James, L.C., Jacques, D.A., Towers, G.J.(2022) Nat Microbiol 7: 1762-1776
- PubMed: 36289397
- DOI: https://doi.org/10.1038/s41564-022-01247-0
- Primary Citation of Related Structures:
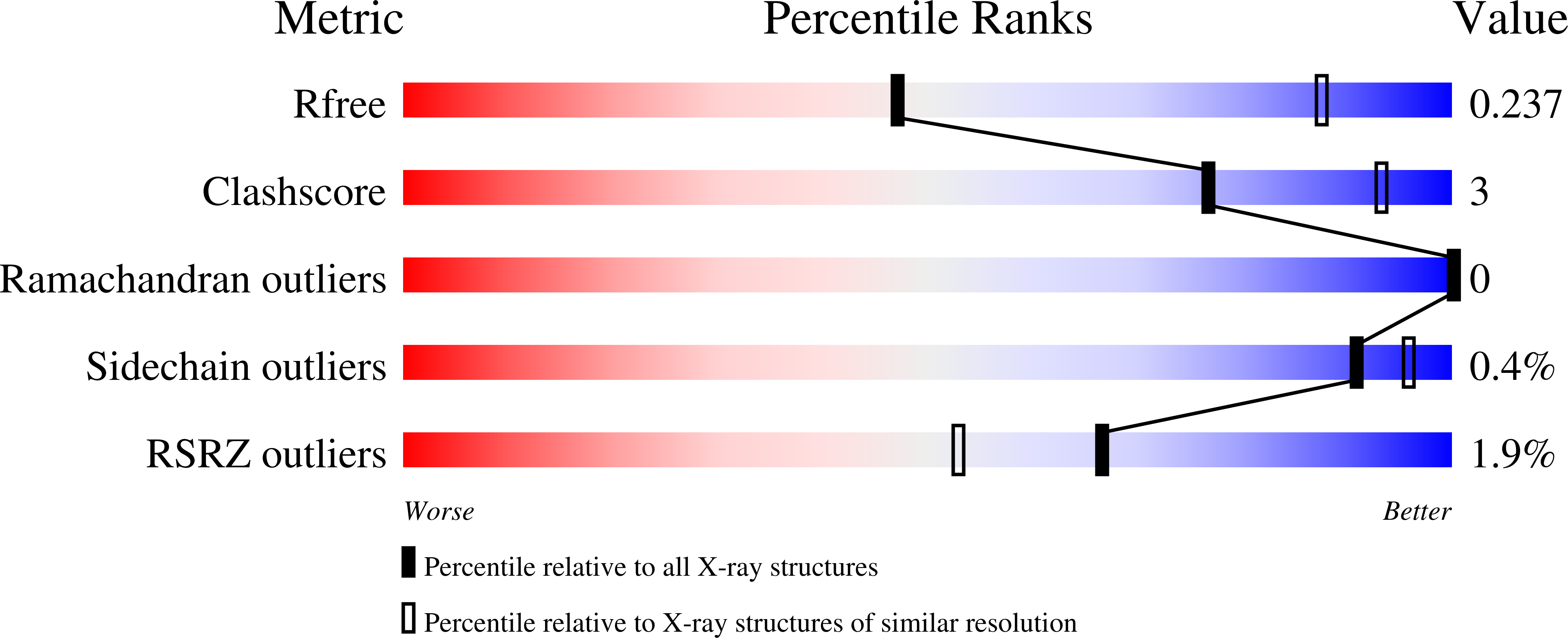
7QDF, 7T12, 7T13, 7T14, 7T15, 8D3B - PubMed Abstract:
Of the 13 known independent zoonoses of simian immunodeficiency viruses to humans, only one, leading to human immunodeficiency virus (HIV) type 1(M) has become pandemic, causing over 80 million human infections. To understand the specific features associated with pandemic human-to-human HIV spread, we compared replication of HIV-1(M) with non-pandemic HIV-(O) and HIV-2 strains in myeloid cell models. We found that non-pandemic HIV lineages replicate less well than HIV-1(M) owing to activation of cGAS and TRIM5-mediated antiviral responses. We applied phylogenetic and X-ray crystallography structural analyses to identify differences between pandemic and non-pandemic HIV capsids. We found that genetic reversal of two specific amino acid adaptations in HIV-1(M) enables activation of TRIM5, cGAS and innate immune responses. We propose a model in which the parental lineage of pandemic HIV-1(M) evolved a capsid that prevents cGAS and TRIM5 triggering, thereby allowing silent replication in myeloid cells. We hypothesize that this capsid adaptation promotes human-to-human spread through avoidance of innate immune response activation.
Organizational Affiliation:
Division of Infection and Immunity, UCL, London, UK.