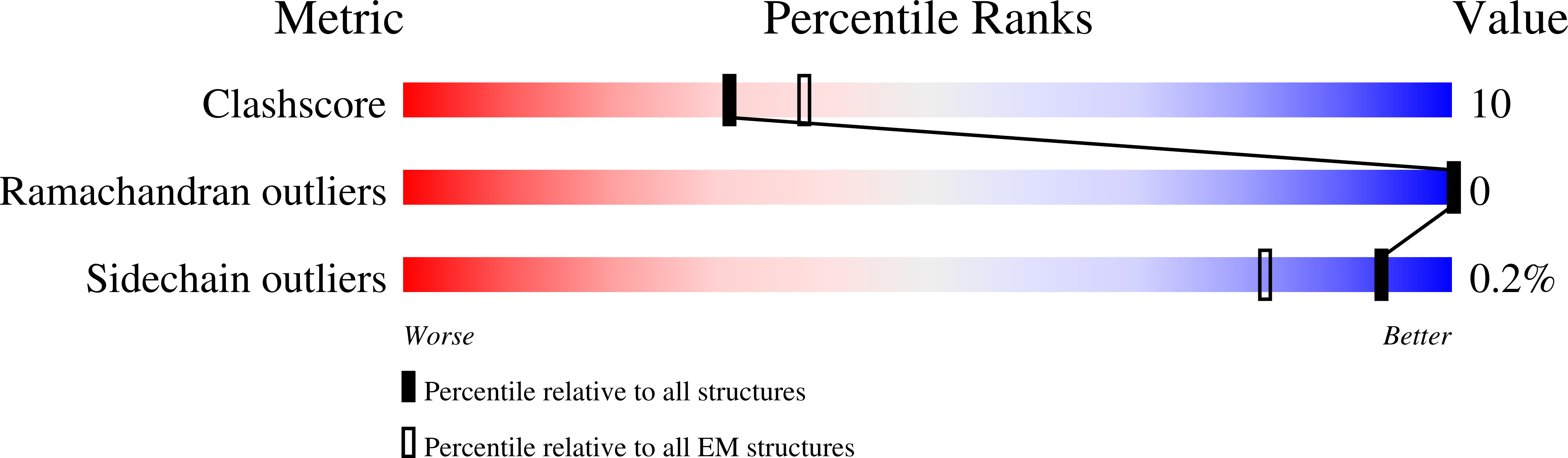Neutralizing antibodies protect mice against Venezuelan equine encephalitis virus aerosol challenge.
Kafai, N.M., Williamson, L.E., Binshtein, E., Sukupolvi-Petty, S., Gardner, C.L., Liu, J., Mackin, S., Kim, A.S., Kose, N., Carnahan, R.H., Jung, A., Droit, L., Reed, D.S., Handley, S.A., Klimstra, W.B., Crowe, J.E., Diamond, M.S.(2022) J Exp Med 219
- PubMed: 35297953
- DOI: https://doi.org/10.1084/jem.20212532
- Primary Citation of Related Structures:
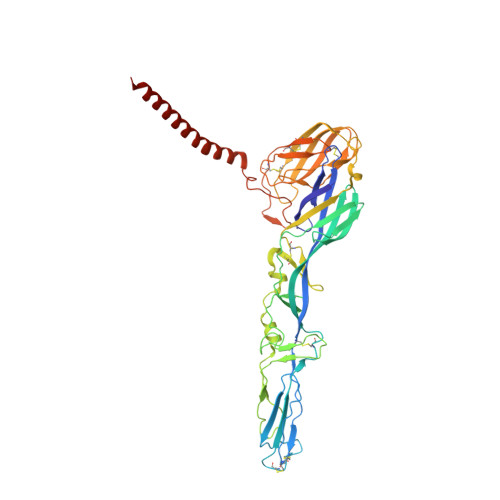
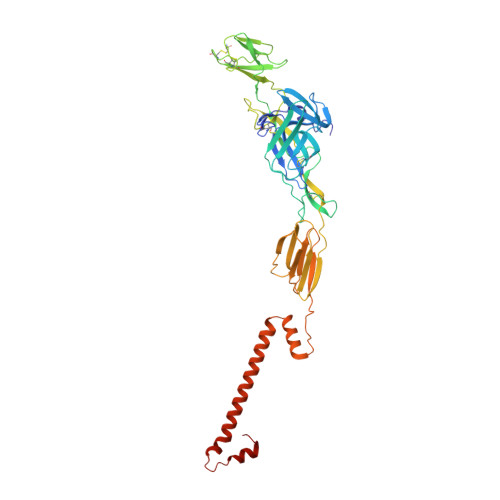
7SFU, 7SFV, 7SFW - PubMed Abstract:
Venezuelan equine encephalitis virus (VEEV) remains a risk for epidemic emergence or use as an aerosolized bioweapon. To develop possible countermeasures, we isolated VEEV-specific neutralizing monoclonal antibodies (mAbs) from mice and a human immunized with attenuated VEEV strains. Functional assays and epitope mapping established that potently inhibitory anti-VEEV mAbs bind distinct antigenic sites in the A or B domains of the E2 glycoprotein and block multiple steps in the viral replication cycle including attachment, fusion, and egress. A 3.2-Å cryo-electron microscopy reconstruction of VEEV virus-like particles bound by a human Fab suggests that antibody engagement of the B domain may result in cross-linking of neighboring spikes to prevent conformational requirements for viral fusion. Prophylaxis or postexposure therapy with these mAbs protected mice against lethal aerosol challenge with VEEV. Our study defines functional and structural mechanisms of mAb protection and suggests that multiple antigenic determinants on VEEV can be targeted for vaccine or antibody-based therapeutic development.
Organizational Affiliation:
Department of Medicine, Washington University School of Medicine, St. Louis, MO.