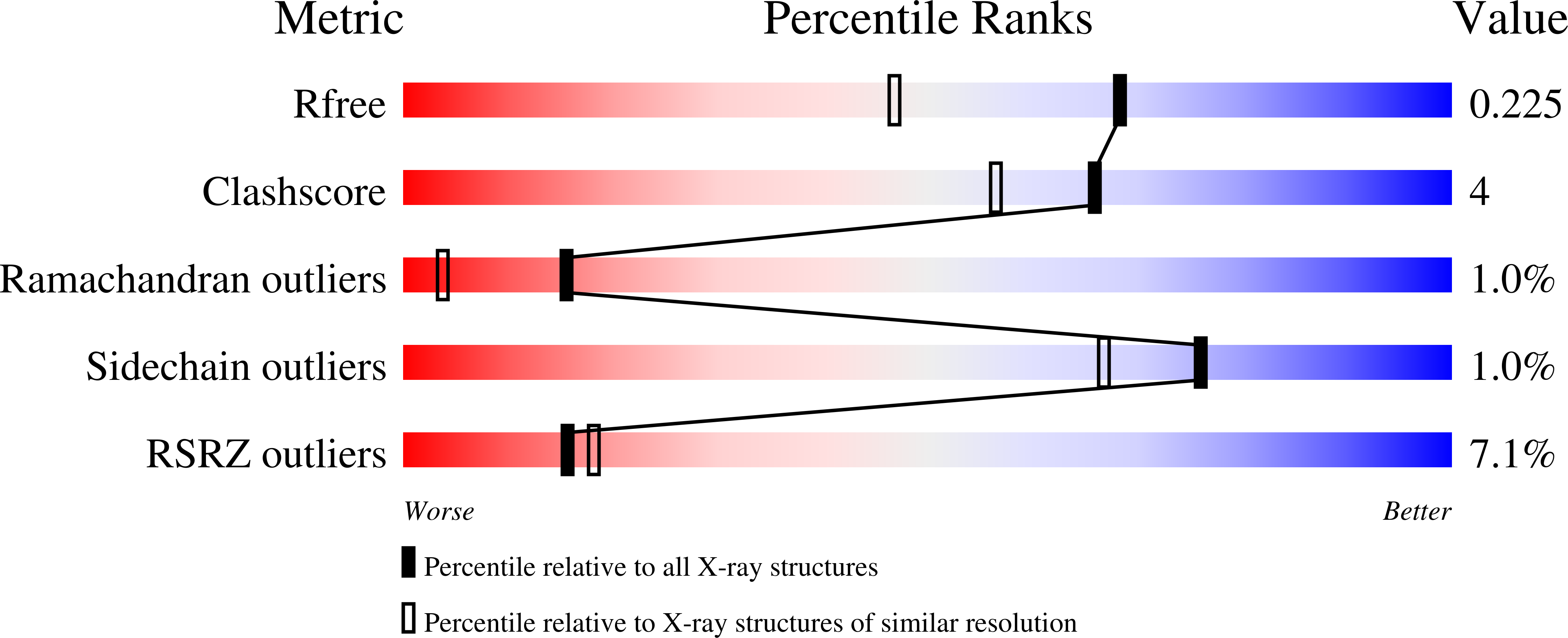
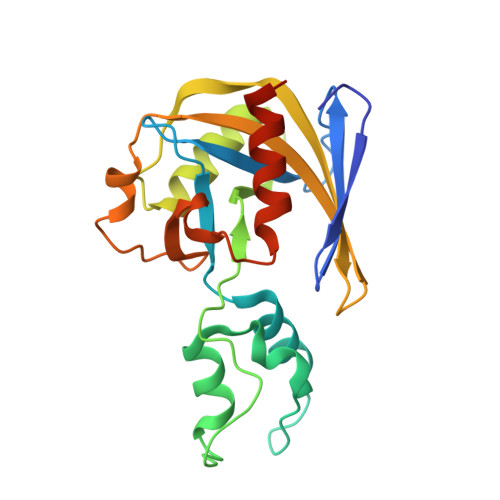
Structure of the poxvirus decapping enzyme D9 reveals its mechanism of cap recognition and catalysis.
Peters, J.K., Tibble, R.W., Warminski, M., Jemielity, J., Gross, J.D.(2022) Structure 30: 721
- PubMed: 35290794
- DOI: https://doi.org/10.1016/j.str.2022.02.012
- Primary Citation of Related Structures:
7SEZ, 7SF0 - PubMed Abstract:
Poxviruses encode decapping enzymes that remove the protective 5' cap from both host and viral mRNAs to commit transcripts for decay by the cellular exonuclease Xrn1. Decapping by these enzymes is critical for poxvirus pathogenicity by means of simultaneously suppressing host protein synthesis and limiting the accumulation of viral double-stranded RNA (dsRNA), a trigger for antiviral responses. Here we present a high-resolution structural view of the vaccinia virus decapping enzyme D9. This Nudix enzyme contains a domain organization different from other decapping enzymes in which a three-helix bundle is inserted into the catalytic Nudix domain. The 5' mRNA cap is positioned in a bipartite active site at the interface of the two domains. Specificity for the methylated guanosine cap is achieved by stacking between conserved aromatic residues in a manner similar to that observed in canonical cap-binding proteins VP39, eIF4E, and CBP20, and distinct from eukaryotic decapping enzyme Dcp2.
Organizational Affiliation:
Department of Pharmaceutical Chemistry, University of California, San Francisco, San Francisco, CA 94158, USA.