Discovery of a new flavin N5-adduct in a tyrosine to phenylalanine variant of d-Arginine dehydrogenase.
Iyer, A., Reis, R.A.G., Agniswamy, J., Weber, I.T., Gadda, G.(2021) Arch Biochem Biophys 715: 109100-109100
- PubMed: 34864048
- DOI: https://doi.org/10.1016/j.abb.2021.109100
- Primary Citation of Related Structures:
7RDF - PubMed Abstract:
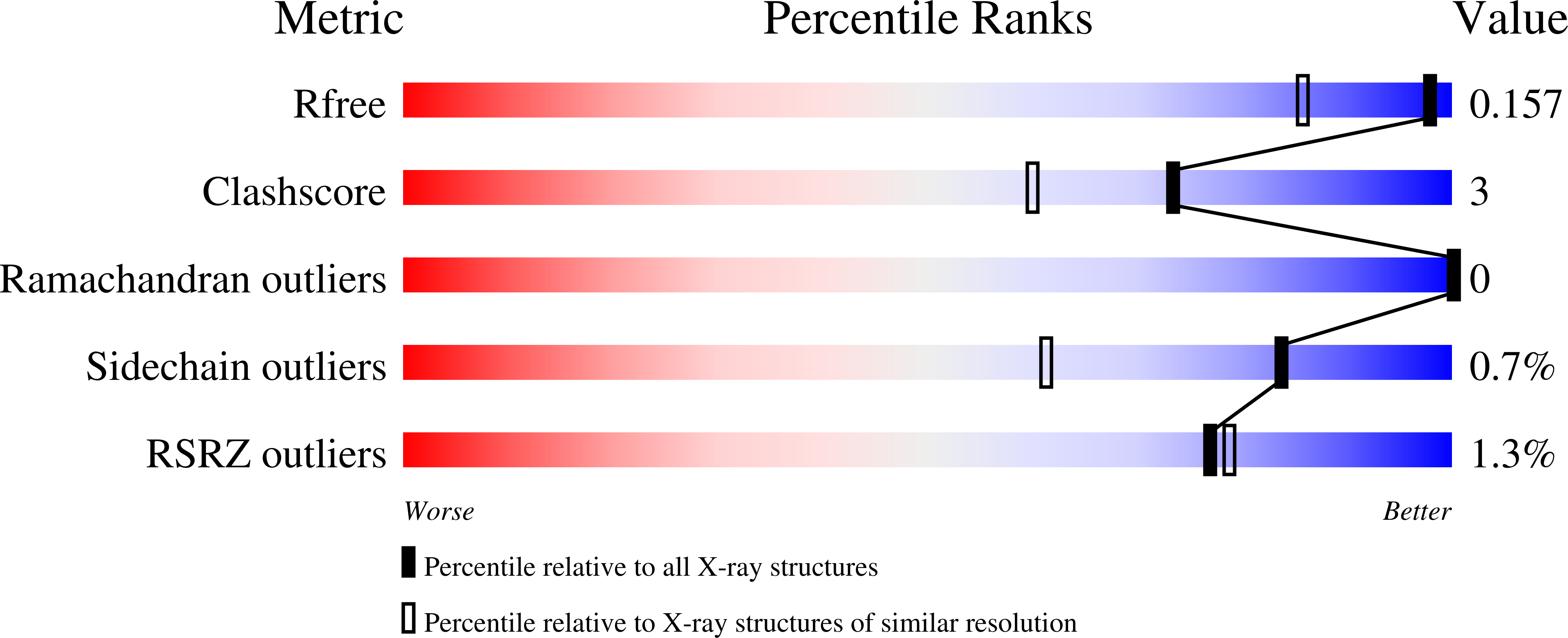
d-Arginine dehydrogenase from Pseudomonas aeruginosa (PaDADH) catalyzes the flavin-dependent oxidation of d-arginine and other d-amino acids. Here, we report the crystal structure at 1.29 Å resolution for PaDADH-Y249F expressed and co-crystallized with d-arginine. The overall structure of PaDADH-Y249F resembled PaDADH-WT, but the electron density for the flavin cofactor was ambiguous, suggesting the presence of modified flavins. Electron density maps and mass spectrometric analysis confirmed the presence of both N5-(4-guanidino-oxobutyl)-FAD and 6-OH-FAD in a single crystal of PaDADH-Y249F and helped with the further refinement of the X-ray crystal structure. The versatility of the reduced flavin is apparent in the PaDADH-Y249F structure and is evidenced by the multiple functions it can perform in the same active site.
Organizational Affiliation:
Department of Chemistry, Georgia State University, Atlanta, GA, 30302, USA.