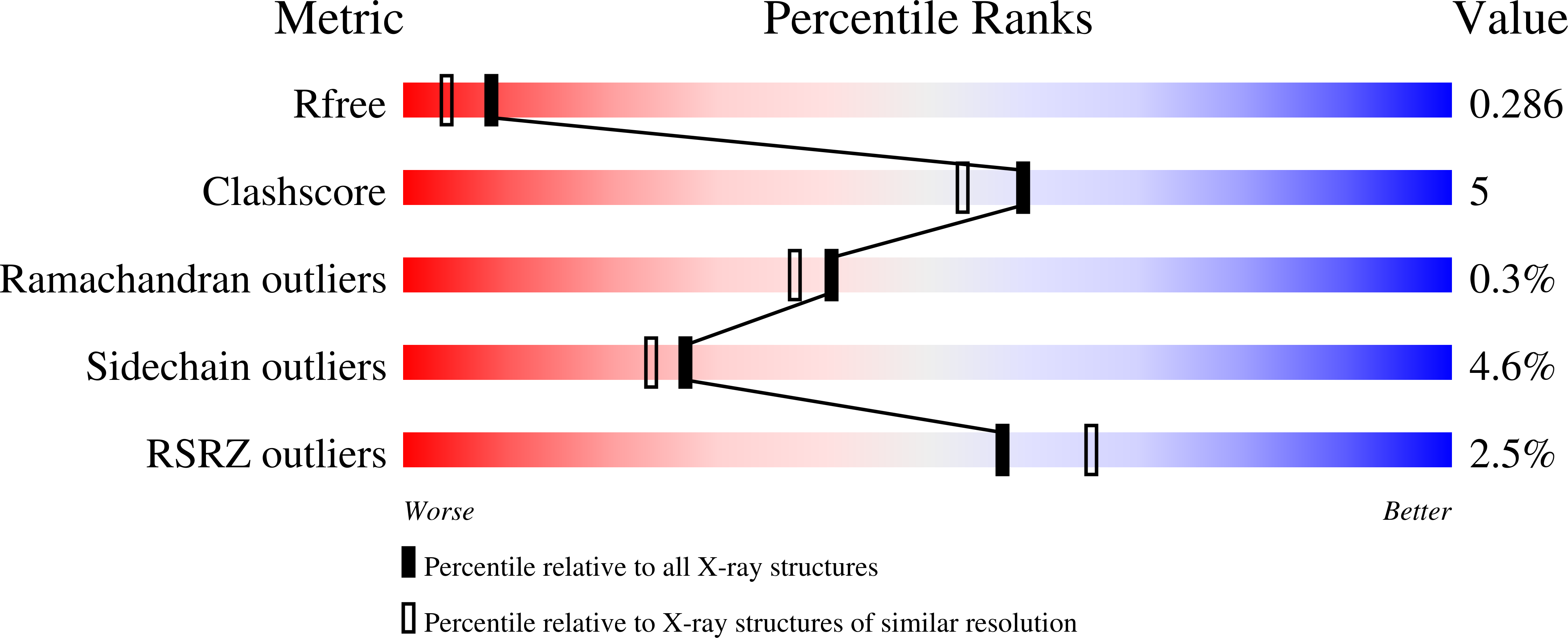
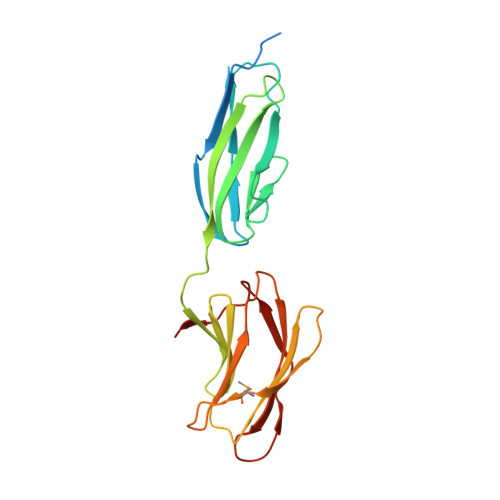
Structural and biochemical characterization of C-terminal Big_2-CBM56 domains of Bacillus circulans IAM1165 beta-1,3-glucanase H and Paenibacillus sp CBM56
Najmudin, S., Venditto, I., Pires, V.R., Caseiro, C., Correia, M.A.S., Romao, M.J., Carvalho, A.L., Fontes, C.M.G.A., Bule, P.To be published.