Family 1 Glycosyltransferase UGT706F8 from Zea mays Selectively Catalyzes the Synthesis of Silibinin 7- O -beta-d-Glucoside.
Bidart, G.N., Putkaradze, N., Fredslund, F., Kjeldsen, C., Ruiz, A.G., Duus, J.O., Teze, D., Welner, D.H.(2022) ACS Sustain Chem Eng 10: 5078-5083
- PubMed: 35493695
- DOI: https://doi.org/10.1021/acssuschemeng.1c07593
- Primary Citation of Related Structures:
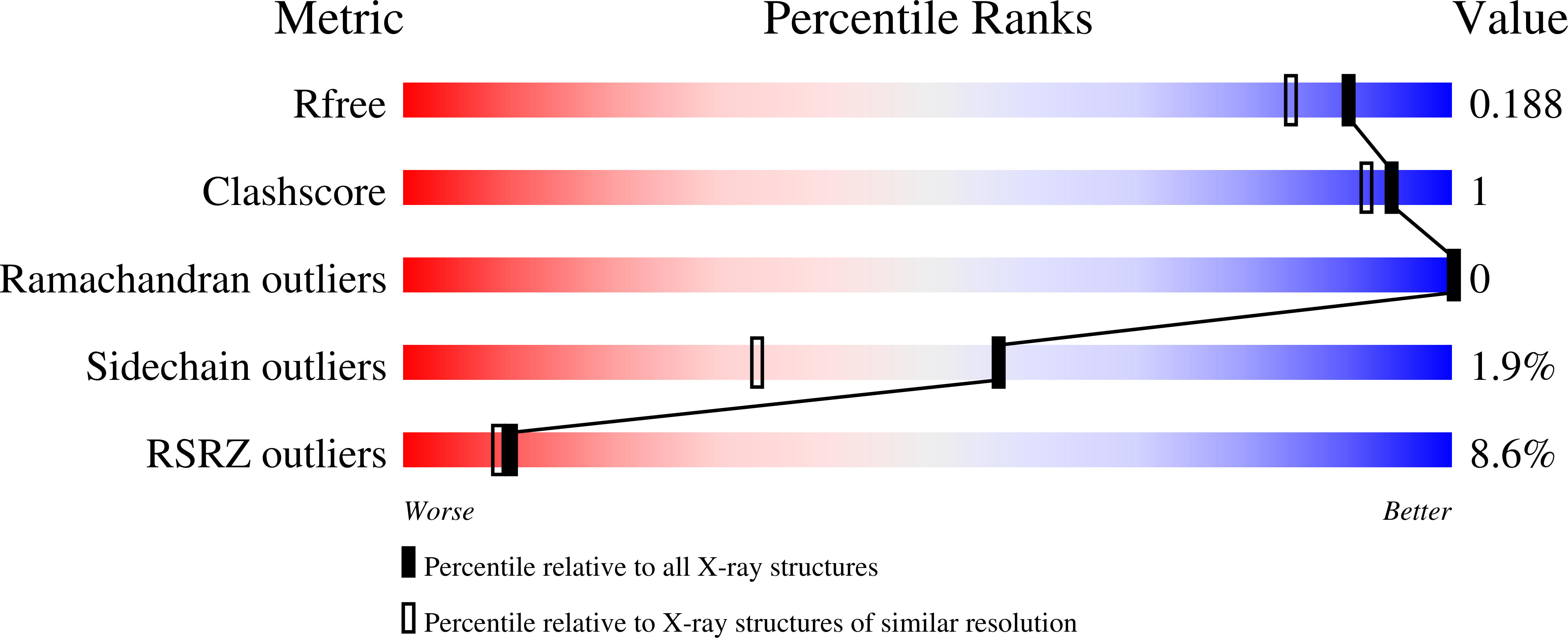
7Q3S - PubMed Abstract:
Regioselective glycosylation is a chemical challenge, leading to multistep syntheses with protecting group manipulations, ultimately resulting in poor atom economy and compromised sustainability. Enzymes allow eco-friendly and regioselective bond formation with fully deprotected substrates in a single reaction. For the selective glucosylation of silibinin, a pharmaceutical challenged with low solubility, enzyme engineering has previously been employed, but the resulting yields and k cat were limited, prohibiting the application of the engineered catalyst. Here, we identified a naturally regioselective silibinin glucosyltransferase, UGT706F8, a family 1 glycosyltransferase from Zea mays . It selectively and efficiently ( k cat = 2.1 ± 0.1 s -1 ; K M = 36.9 ± 5.2 μM; TTN = 768 ± 22) catalyzes the quantitative synthesis of silibinin 7- O -β-d-glucoside. We solved the crystal structure of UGT706F8 and investigated the molecular determinants of regioselective silibinin glucosylation. UGT706F8 was the only regioselective enzyme among 18 glycosyltransferases found to be active on silibinin. We found the temperature optimum of UGT706F8 to be 34 °C and the pH optimum to be 7-8. Our results indicate that UGT706F8 is an efficient silibinin glycosyltransferase that enables biocatalytic production of silbinin 7- O -β-d-glucoside.
Organizational Affiliation:
The Novo Nordisk Foundation Center for Biosustainability, Technical University of Denmark, DK-2800 Kongens Lyngby, Denmark.