Agaricales Mushroom Lignin Peroxidase: From Structure-Function to Degradative Capabilities.
Sanchez-Ruiz, M.I., Ayuso-Fernandez, I., Rencoret, J., Gonzalez-Ramirez, A.M., Linde, D., Davo-Siguero, I., Romero, A., Gutierrez, A., Martinez, A.T., Ruiz-Duenas, F.J.(2021) Antioxidants (Basel) 10
- PubMed: 34573078
- DOI: https://doi.org/10.3390/antiox10091446
- Primary Citation of Related Structures:
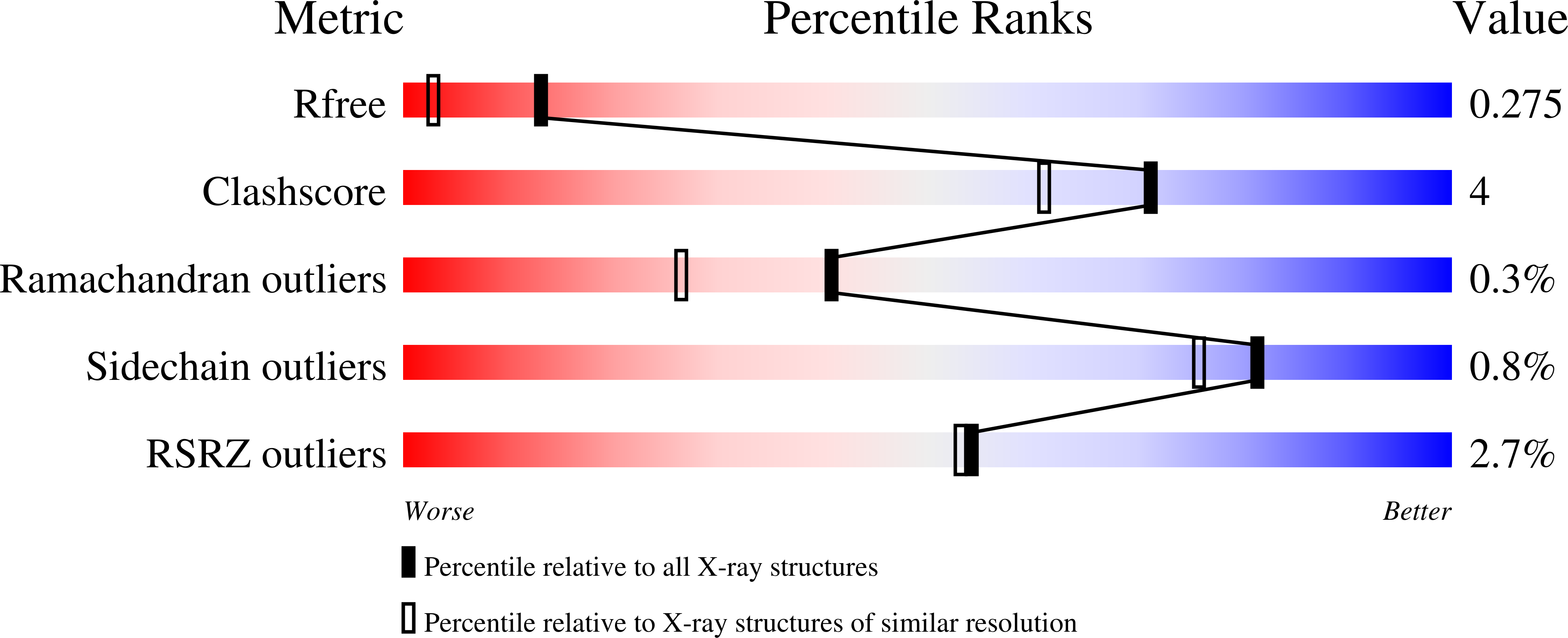
7OO5 - PubMed Abstract:
Lignin biodegradation has been extensively studied in white-rot fungi, which largely belong to order Polyporales. Among the enzymes that wood-rotting polypores secrete, lignin peroxidases (LiPs) have been labeled as the most efficient. Here, we characterize a similar enzyme (ApeLiP) from a fungus of the order Agaricales (with ~13,000 described species), the soil-inhabiting mushroom Agrocybe pediades . X-ray crystallography revealed that ApeLiP is structurally related to Polyporales LiPs, with a conserved heme-pocket and a solvent-exposed tryptophan. Its biochemical characterization shows that ApeLiP can oxidize both phenolic and non-phenolic lignin model-compounds, as well as different dyes. Moreover, using stopped-flow rapid spectrophotometry and 2D-NMR, we demonstrate that ApeLiP can also act on real lignin. Characterization of a variant lacking the above tryptophan residue shows that this is the oxidation site for lignin and other high redox-potential substrates, and also plays a role in phenolic substrate oxidation. The reduction potentials of the catalytic-cycle intermediates were estimated by stopped-flow in equilibrium reactions, showing similar activation by H 2 O 2 , but a lower potential for the rate-limiting step (compound-II reduction) compared to other LiPs. Unexpectedly, ApeLiP was stable from acidic to basic pH, a relevant feature for application considering its different optima for oxidation of phenolic and nonphenolic compounds.
Organizational Affiliation:
Centro de Investigaciones Biológicas "Margarita Salas" (CIB), Consejo Superior de Investigaciones Científicas (CSIC), 28040 Madrid, Spain.