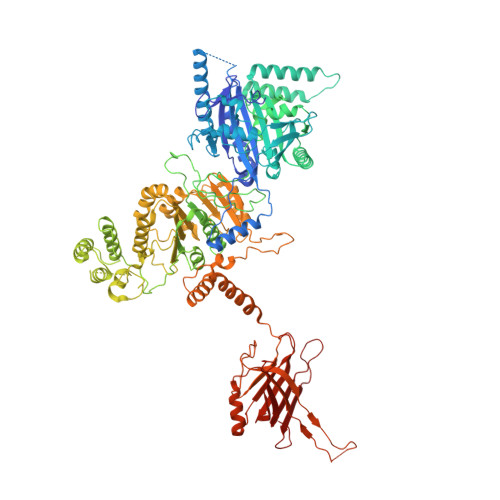
Cryo-EM structure of a type IV secretion system.
Mace, K., Vadakkepat, A.K., Redzej, A., Lukoyanova, N., Oomen, C., Braun, N., Ukleja, M., Lu, F., Costa, T.R.D., Orlova, E.V., Baker, D., Cong, Q., Waksman, G.(2022) Nature 607: 191-196
- PubMed: 35732732
- DOI: https://doi.org/10.1038/s41586-022-04859-y
- Primary Citation of Related Structures:
7O3J, 7O3T, 7O3V, 7O41, 7O42, 7O43, 7OIU, 7Q1V - PubMed Abstract:
Bacterial conjugation is the fundamental process of unidirectional transfer of DNAs, often plasmid DNAs, from a donor cell to a recipient cell 1 . It is the primary means by which antibiotic resistance genes spread among bacterial populations 2,3 . In Gram-negative bacteria, conjugation is mediated by a large transport apparatus-the conjugative type IV secretion system (T4SS)-produced by the donor cell and embedded in both its outer and inner membranes. The T4SS also elaborates a long extracellular filament-the conjugative pilus-that is essential for DNA transfer 4,5 . Here we present a high-resolution cryo-electron microscopy (cryo-EM) structure of a 2.8 megadalton T4SS complex composed of 92 polypeptides representing 8 of the 10 essential T4SS components involved in pilus biogenesis. We added the two remaining components to the structural model using co-evolution analysis of protein interfaces, to enable the reconstitution of the entire system including the pilus. This structure describes the exceptionally large protein-protein interaction network required to assemble the many components that constitute a T4SS and provides insights on the unique mechanism by which they elaborate pili.
Organizational Affiliation:
Institute of Structural and Molecular Biology, Department of Biological Sciences, Birkbeck College, London, UK. k.mace@mail.cryst.bbk.ac.uk.