Crystallographic structure of human Tsg101 UEV domain in complex with a HEV ORF3 peptide
Moschidi, D., Dupre, E., Villeret, V., Hanoulle, X.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tsg101 UEV domain | 147 | Homo sapiens | Mutation(s): 0 Gene Names: TSG101 | 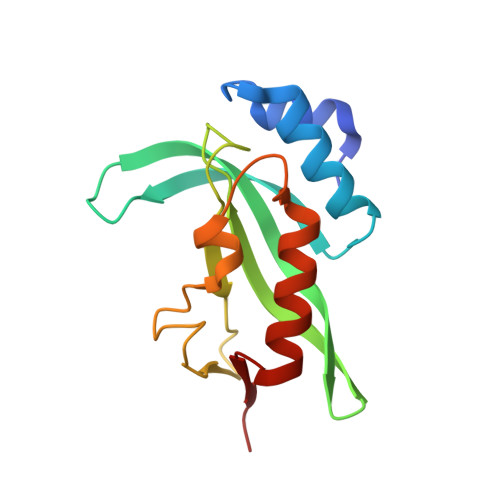 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q99816 (Homo sapiens) Explore Q99816 Go to UniProtKB: Q99816 | |||||
PHAROS: Q99816 GTEx: ENSG00000074319 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q99816 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein ORF3 | 10 | Paslahepevirus balayani | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for Q9YLR0 (Hepatitis E virus genotype 3 (isolate Human/United States/US2)) Explore Q9YLR0 Go to UniProtKB: Q9YLR0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9YLR0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SO4 Query on SO4 | E [auth A] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| CL Query on CL | C [auth A], D [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| NH4 Query on NH4 | AA [auth A] AB [auth B] BA [auth A] BB [auth B] CA [auth A] | AMMONIUM ION H4 N QGZKDVFQNNGYKY-UHFFFAOYSA-O |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 42.57 | α = 90 |
| b = 42.57 | β = 90 |
| c = 188.16 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| XDS | data reduction |
| Coot | model building |
| Aimless | data scaling |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Agence Nationale de Recherches Sur le Sida et les Hepatites Virales (ANRS) | France | ECTZ101316 |