Crystal structure of SU3327 (halicin) covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2.
Costanzi, E., Demitri, N., Giabbai, B., Storici, P.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Main Protease | 306 | Severe acute respiratory syndrome coronavirus 2 | Mutation(s): 0 Gene Names: rep, 1a-1b EC: 3.4.19.12 (PDB Primary Data), 3.4.22 (PDB Primary Data), 3.4.22.69 (PDB Primary Data), 2.7.7.48 (PDB Primary Data), 3.6.4.12 (PDB Primary Data), 3.6.4.13 (PDB Primary Data), 3.1.13 (PDB Primary Data), 3.1 (PDB Primary Data), 2.1.1 (PDB Primary Data) | 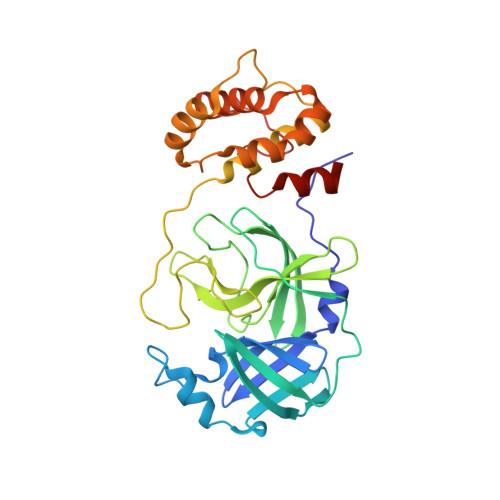 | |
UniProt | |||||
Find proteins for P0DTD1 (Severe acute respiratory syndrome coronavirus 2) Explore P0DTD1 Go to UniProtKB: P0DTD1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0DTD1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| U88 (Subject of Investigation/LOI) Query on U88 | C [auth A] D [auth A] E [auth A] H [auth A] J [auth B] | 5-nitro-1,3-thiazole C3 H2 N2 O2 S VVVCJCRUFSIVHI-UHFFFAOYSA-N |  | ||
| SO4 Query on SO4 | M [auth B] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| NO3 Query on NO3 | F [auth A], G [auth A], N [auth B] | NITRATE ION N O3 NHNBFGGVMKEFGY-UHFFFAOYSA-N |  | ||
| CL Query on CL | I [auth A], Q [auth B] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| NA Query on NA | P [auth B] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 68.234 | α = 90 |
| b = 100.015 | β = 90 |
| c = 103.698 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| European Commission | European Union | 101003551 |