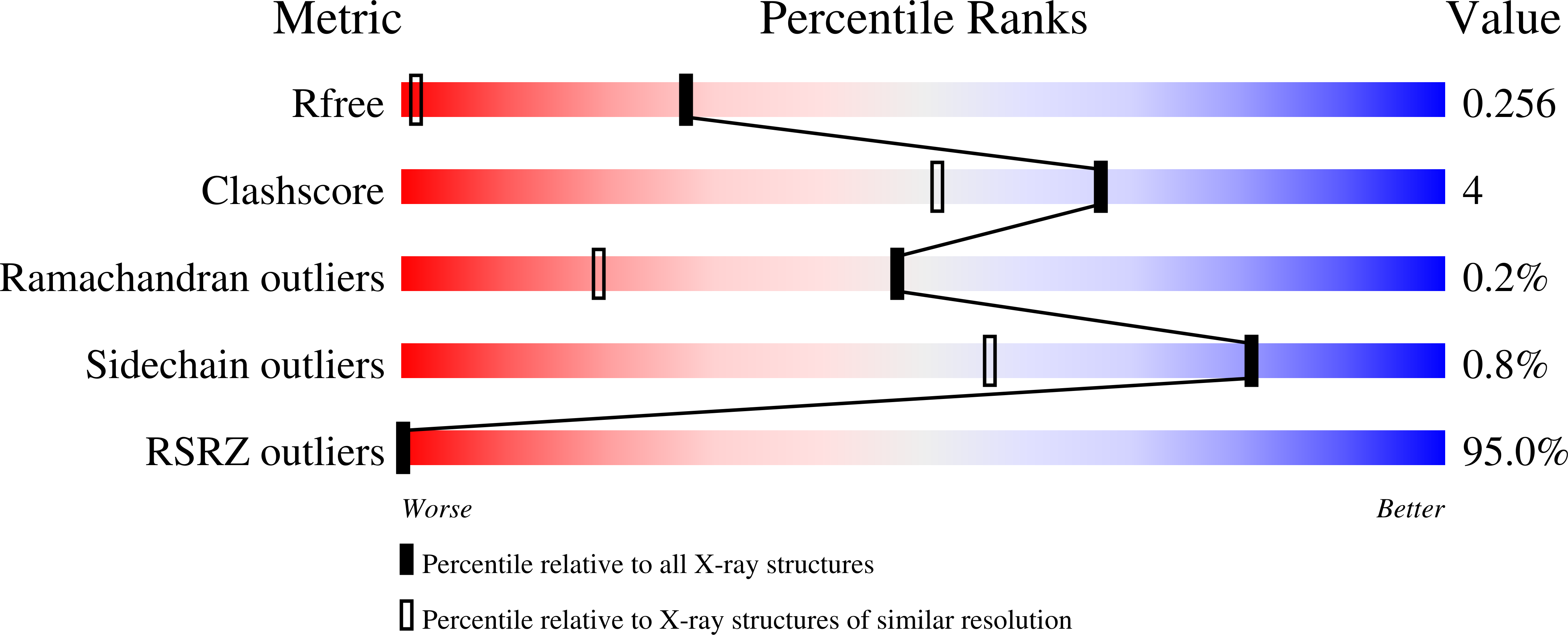
The first crystal structure of a xylobiose-bound xylobiohydrolase with high functional specificity from the bacterial glycoside hydrolase family 30, subfamily 10.
St John, F.J., Crooks, C., Kim, Y., Tan, K., Joachimiak, A.(2022) FEBS Lett 596: 2449-2464
- PubMed: 35876256
- DOI: https://doi.org/10.1002/1873-3468.14454
- Primary Citation of Related Structures:
7N6H, 7N6O - PubMed Abstract:
Xylobiose is a prebiotic sugar that has applications in functional foods. This report describes the first X-ray crystallographic structure models of apo and xylobiose-bound forms of a xylobiohydrolase (XBH) from Acetivibrio clariflavus. This xylan-active enzyme, a member of the recently described glycoside hydrolase family 30 (GH30), subfamily 10, phylogenetic clade has been shown to strictly release xylobiose as its primary hydrolysis product. Inspection of the apo structure reveals a glycone region X 2 -binding slot. When X 2 binds, the non-reducing xylose in the -2 subsite is highly coordinated with numerous hydrogen bond contacts while contacts in the -1 subsite mostly reflect interactions typical for GH30 and enzymes in clan A of the carbohydrate-active enzymes database (CAZy). This structure provides an explanation for the high functional specificity of this new bacterial GH30 XBH subfamily.
Organizational Affiliation:
Institute for Microbial and Biochemical Technology, Forest Products Laboratory, USDA Forest Service, Madison, WI, USA.