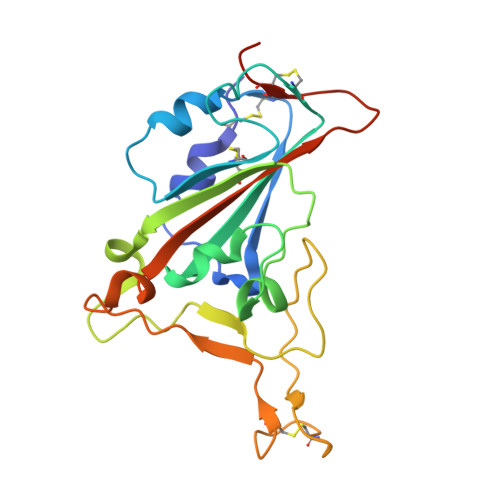
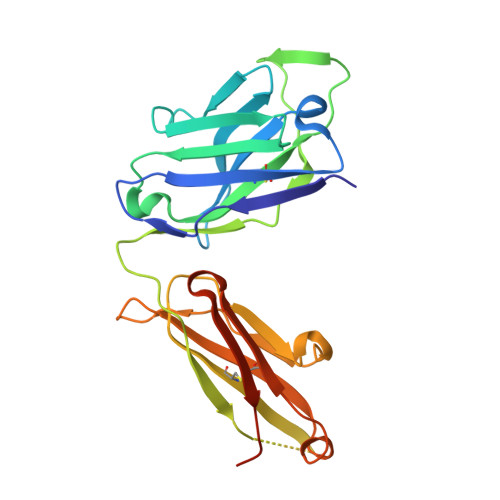
Immunizations with diverse sarbecovirus receptor-binding domains elicit SARS-CoV-2 neutralizing antibodies against a conserved site of vulnerability.
Burnett, D.L., Jackson, K.J.L., Langley, D.B., Aggrawal, A., Stella, A.O., Johansen, M.D., Balachandran, H., Lenthall, H., Rouet, R., Walker, G., Saunders, B.M., Singh, M., Li, H., Henry, J.Y., Jackson, J., Stewart, A.G., Witthauer, F., Spence, M.A., Hansbro, N.G., Jackson, C., Schofield, P., Milthorpe, C., Martinello, M., Schulz, S.R., Roth, E., Kelleher, A., Emery, S., Britton, W.J., Rawlinson, W.D., Karl, R., Schafer, S., Winkler, T.H., Brink, R., Bull, R.A., Hansbro, P.M., Jack, H.M., Turville, S., Christ, D., Goodnow, C.C.(2021) Immunity 54: 2908-2921.e6
- PubMed: 34788600
- DOI: https://doi.org/10.1016/j.immuni.2021.10.019
- Primary Citation of Related Structures:
7MSQ - PubMed Abstract:
Viral mutations are an emerging concern in reducing SARS-CoV-2 vaccination efficacy. Second-generation vaccines will need to elicit neutralizing antibodies against sites that are evolutionarily conserved across the sarbecovirus subgenus. Here, we immunized mice containing a human antibody repertoire with diverse sarbecovirus receptor-binding domains (RBDs) to identify antibodies targeting conserved sites of vulnerability. Antibodies with broad reactivity against diverse clade B RBDs targeting the conserved class 4 epitope, with recurring IGHV/IGKV pairs, were readily elicited but were non-neutralizing. However, rare class 4 antibodies binding this conserved RBD supersite showed potent neutralization of SARS-CoV-2 and all variants of concern. Structural analysis revealed that the neutralizing ability of cross-reactive antibodies was reserved only for those with an elongated CDRH3 that extends the antiparallel beta-sheet RBD core and orients the antibody light chain to obstruct ACE2-RBD interactions. These results identify a structurally defined pathway for vaccine strategies eliciting escape-resistant SARS-CoV-2 neutralizing antibodies.
Organizational Affiliation:
Garvan Institute of Medical Research, Sydney, NSW 2010, Australia; UNSW Sydney, Faculty of Medicine, Sydney, NSW 2010, Australia. Electronic address: d.burnett@garvan.org.au.