The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with ebselen
Osipiuk, J., Tesar, C., Endres, M., Maltseva, N., Joachimiak, A., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Papain-like protease | 318 | Severe acute respiratory syndrome coronavirus 2 | Mutation(s): 1 EC: 3.4.22 | 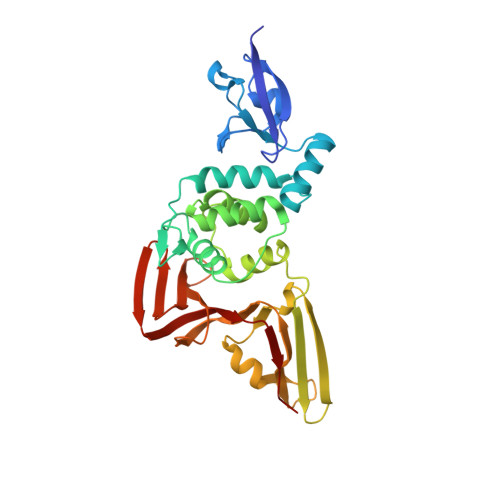 | |
UniProt | |||||
Find proteins for P0DTD1 (Severe acute respiratory syndrome coronavirus 2) Explore P0DTD1 Go to UniProtKB: P0DTD1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0DTD1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 7 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 9JT (Subject of Investigation/LOI) Query on 9JT | C [auth A], S [auth B] | N-phenyl-2-selanylbenzamide C13 H11 N O Se PVPUYGNPKBMXGO-UHFFFAOYSA-N |  | ||
| IOD Query on IOD | AA [auth B] G [auth A] H [auth A] I [auth A] J [auth A] | IODIDE ION I XMBWDFGMSWQBCA-UHFFFAOYSA-M |  | ||
| GOL Query on GOL | F [auth A], V [auth B] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| ZN Query on ZN | D [auth A], T [auth B] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| FMT Query on FMT | BA [auth B], M [auth A], N [auth A], O [auth A] | FORMIC ACID C H2 O2 BDAGIHXWWSANSR-UHFFFAOYSA-N |  | ||
| CL Query on CL | E [auth A], U [auth B] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| NA Query on NA | CA [auth B], P [auth A] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 114.887 | α = 90 |
| b = 114.887 | β = 90 |
| c = 252.82 | γ = 120 |
| Software Name | Purpose |
|---|---|
| HKL-3000 | data scaling |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| HKL-3000 | data reduction |
| HKL-3000 | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | United States | HHSN272201200026C |
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | United States | HHSN272201700060C |