To be provided
Aleshin, A.E., Shiryaev, S.A., Liddington, R.C.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Core protein | 221 | Zika virus | Mutation(s): 0 EC: 3.4.21.91 (PDB Primary Data), 3.6.1.15 (PDB Primary Data), 3.6.4.13 (PDB Primary Data) | 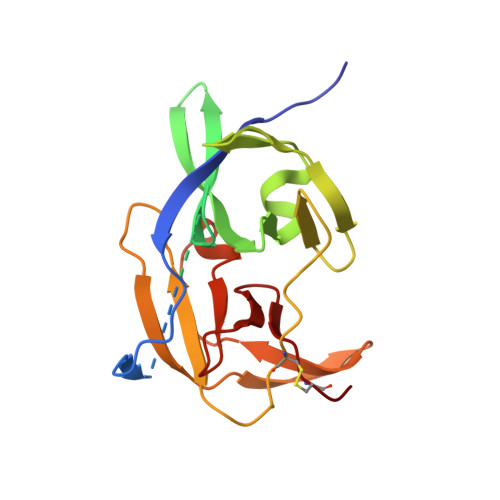 | |
UniProt | |||||
Find proteins for A0A024B7W1 (Zika virus (isolate ZIKV/Human/French Polynesia/10087PF/2013)) Explore A0A024B7W1 Go to UniProtKB: A0A024B7W1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A024B7W1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| YO1 (Subject of Investigation/LOI) Query on YO1 | C [auth A] | 4-(2-{2,4-diamino-5-[2-(4-{[(2E)-1,3-thiazolidin-2-ylidene]sulfamoyl}phenyl)hydrazinyl]phenyl}hydrazinyl)-N-[(2S)-1,3-thiazolidin-2-yl]benzene-1-sulfonamide C24 H20 N10 O4 S4 BZLWMRWTRRMORD-JWTBXLROSA-N |  | ||
| DMS Query on DMS | G [auth B] | DIMETHYL SULFOXIDE C2 H6 O S IAZDPXIOMUYVGZ-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | D [auth A], F [auth B], H [auth B] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| CL Query on CL | E [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 40.81 | α = 98.88 |
| b = 42.637 | β = 92.11 |
| c = 46.296 | γ = 90.41 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of Dental and Craniofacial Research (NIH/NIDCR) | United States | 5R21AI134581 |