Structural delineation and phase-dependent activation of the costimulatory CD27:CD70 complex.
Liu, W., Maben, Z., Wang, C., Lindquist, K.C., Li, M., Rayannavar, V., Lopez Armenta, I., Nager, A., Pascua, E., Dominik, P.K., Oyen, D., Wang, H., Roach, R.C., Allan, C.M., Mosyak, L., Chaparro-Riggers, J.(2021) J Biol Chem 297: 101102-101102
- PubMed: 34419446
- DOI: https://doi.org/10.1016/j.jbc.2021.101102
- Primary Citation of Related Structures:
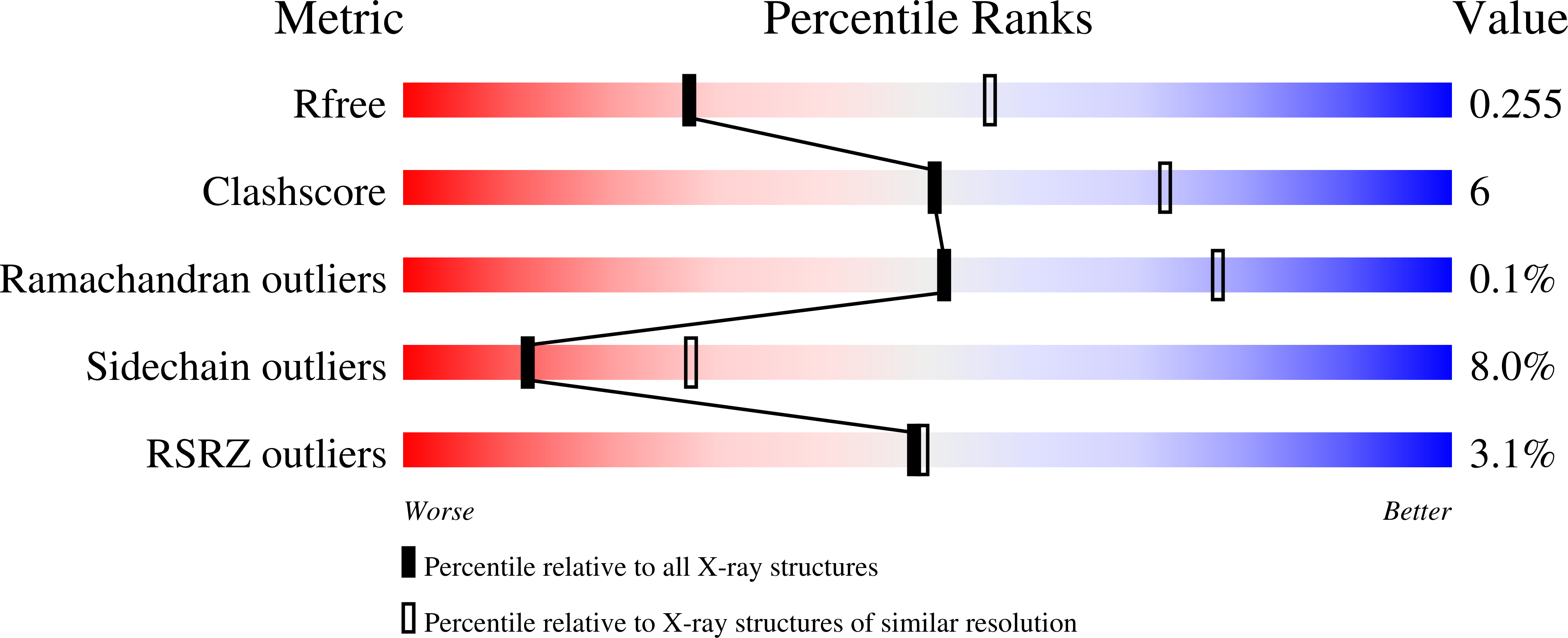
7KX0 - PubMed Abstract:
CD27 is a tumor necrosis factor (TNF) receptor, which stimulates lymphocytes and promotes their differentiation upon activation by TNF ligand CD70. Activation of the CD27 receptor provides a costimulatory signal to promote T cell, B cell, and NK cell activity to facilitate antitumor and anti-infection immunity. Aberrant increased and focused expression of CD70 on many tumor cells renders CD70 an attractive therapeutic target for direct tumor killing. However, despite their use as drug targets to treat cancers, the molecular basis and atomic details of CD27 and CD70 interaction remain elusive. Here we report the crystal structure of human CD27 in complex with human CD70. Analysis of our structure shows that CD70 adopts a classical TNF ligand homotrimeric assembly to engage CD27 receptors in a 3:3 stoichiometry. By combining structural and rational mutagenesis data with reported disease-correlated mutations, we identified the key amino acid residues of CD27 and CD70 that control this interaction. We also report increased potency for plate-bound CD70 constructs compared with solution-phase ligand in a functional activity to stimulate T-cells in vitro. These findings offer new mechanistic insight into this critical costimulatory interaction.
Organizational Affiliation:
Pfizer, Inc, La Jolla, California, USA.