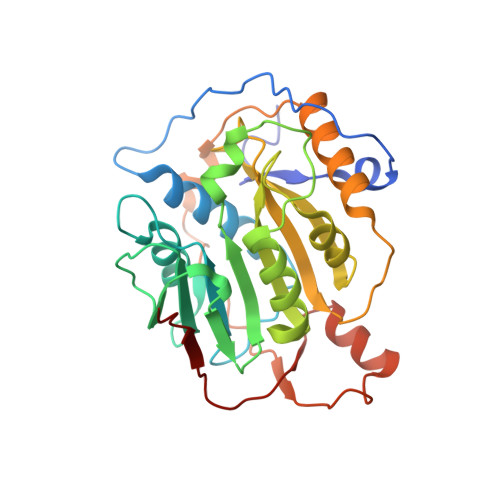
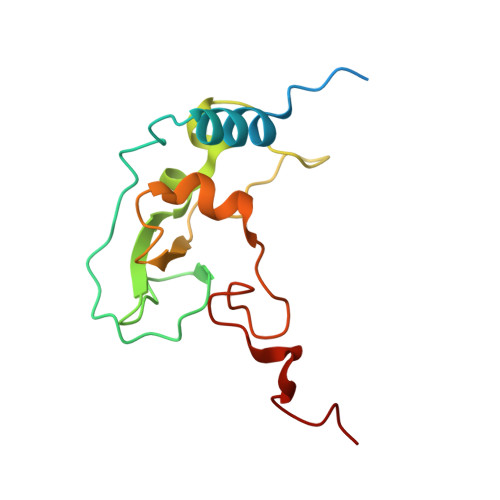
Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with Cap-0 and SAM Determined by Pink-Beam Serial Crystallography
Wilamowski, M., Sherrell, D.A., Minasov, G., Shuvalova, L., Lavens, A., Henning, R., Maltseva, N., Rosas-Lemus, M., Kim, Y., Satchell, K.J.F., Srajer, V., Joachimiak, A., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.