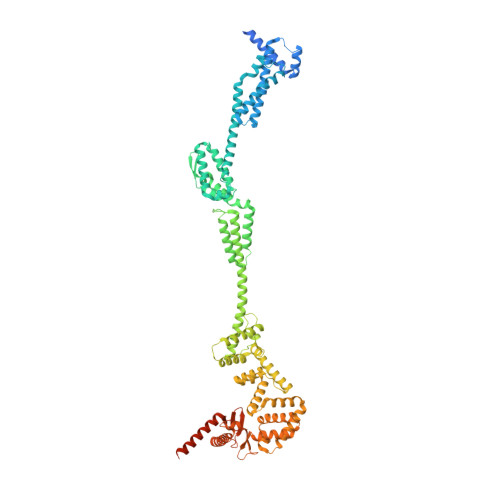
Cryo-EM structure of the periplasmic tunnel of T7 DNA-ejectosome at 2.7 angstrom resolution.
Swanson, N.A., Lokareddy, R.K., Li, F., Hou, C.D., Leptihn, S., Pavlenok, M., Niederweis, M., Pumroy, R.A., Moiseenkova-Bell, V.Y., Cingolani, G.(2021) Mol Cell 81: 3145
- PubMed: 34214465
- DOI: https://doi.org/10.1016/j.molcel.2021.06.001
- Primary Citation of Related Structures:
7K5C - PubMed Abstract:
Hershey and Chase used bacteriophage T2 genome delivery inside Escherichia coli to demonstrate that DNA, not protein, is the genetic material. Seventy years later, our understanding of viral genome delivery in prokaryotes remains limited, especially for short-tailed phages of the Podoviridae family. These viruses expel mysterious ejection proteins found inside the capsid to form a DNA-ejectosome for genome delivery into bacteria. Here, we reconstitute the phage T7 DNA-ejectosome components gp14, gp15, and gp16 and solve the periplasmic tunnel structure at 2.7 Å resolution. We find that gp14 forms an outer membrane pore, gp15 assembles into a 210 Å hexameric DNA tube spanning the host periplasm, and gp16 extends into the host cytoplasm forming a ∼4,200 residue hub. Gp16 promotes gp15 oligomerization, coordinating peptidoglycan hydrolysis, DNA binding, and lipid insertion. The reconstituted gp15:gp16 complex lacks channel-forming activity, suggesting that the pore for DNA passage forms only transiently during genome ejection.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Thomas Jefferson University, 1020 Locust Street, Philadelphia, PA 19107, USA.