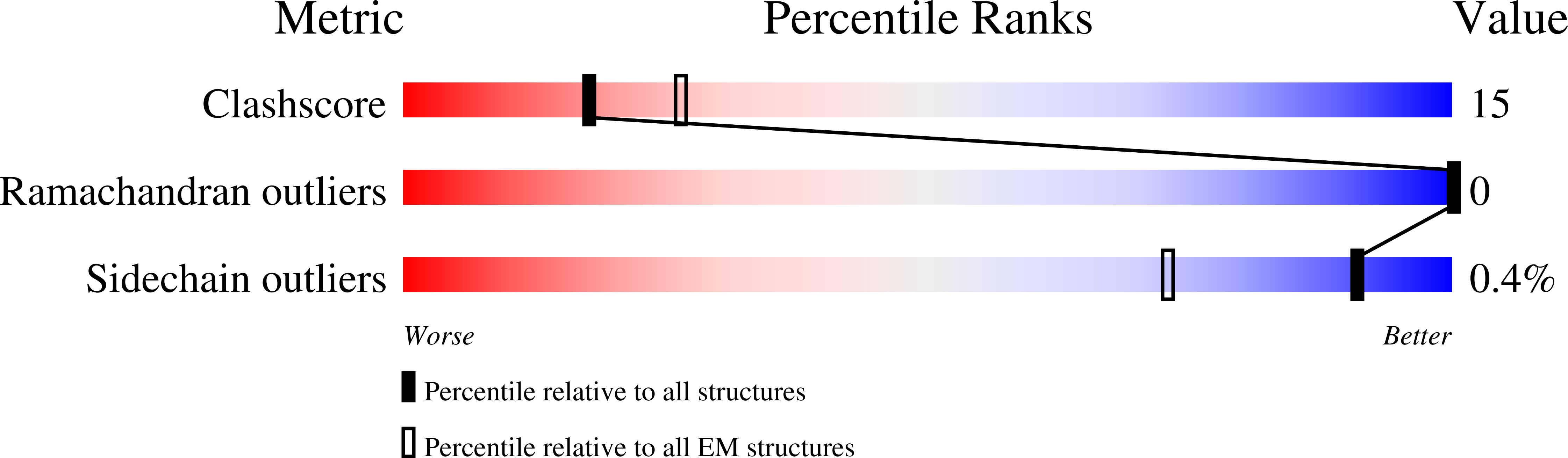The N-terminus of varicella-zoster virus glycoprotein B has a functional role in fusion.
Oliver, S.L., Xing, Y., Chen, D.H., Roh, S.H., Pintilie, G.D., Bushnell, D.A., Sommer, M.H., Yang, E., Carfi, A., Chiu, W., Arvin, A.M.(2021) PLoS Pathog 17: e1008961-e1008961
- PubMed: 33411789
- DOI: https://doi.org/10.1371/journal.ppat.1008961
- Primary Citation of Related Structures:
7K1S - PubMed Abstract:
Varicella-zoster virus (VZV) is a medically important alphaherpesvirus that induces fusion of the virion envelope and the cell membrane during entry, and between cells to form polykaryocytes within infected tissues during pathogenesis. All members of the Herpesviridae, including VZV, have a conserved core fusion complex composed of glycoproteins, gB, gH and gL. The ectodomain of the primary fusogen, gB, has five domains, DI-V, of which DI contains the fusion loops needed for fusion function. We recently demonstrated that DIV is critical for fusion initiation, which was revealed by a 2.8Å structure of a VZV neutralizing mAb, 93k, bound to gB and mutagenesis of the gB-93k interface. To further assess the mechanism of mAb 93k neutralization, the binding site of a non-neutralizing mAb to gB, SG2, was compared to mAb 93k using single particle cryogenic electron microscopy (cryo-EM). The gB-SG2 interface partially overlapped with that of gB-93k but, unlike mAb 93k, mAb SG2 did not interact with the gB N-terminus, suggesting a potential role for the gB N-terminus in membrane fusion. The gB ectodomain structure in the absence of antibody was defined at near atomic resolution by single particle cryo-EM (3.9Å) of native, full-length gB purified from infected cells and by X-ray crystallography (2.4Å) of the transiently expressed ectodomain. Both structures revealed that the VZV gB N-terminus (aa72-114) was flexible based on the absence of visible structures in the cryo-EM or X-ray crystallography data but the presence of gB N-terminal peptides were confirmed by mass spectrometry. Notably, N-terminal residues 109KSQD112 were predicted to form a small α-helix and alanine substitution of these residues abolished cell-cell fusion in a virus-free assay. Importantly, transferring the 109AAAA112 mutation into the VZV genome significantly impaired viral propagation. These data establish a functional role for the gB N-terminus in membrane fusion broadly relevant to the Herpesviridae.
Organizational Affiliation:
Department of Pediatrics, Stanford University School of Medicine, Stanford, California, United States of America.