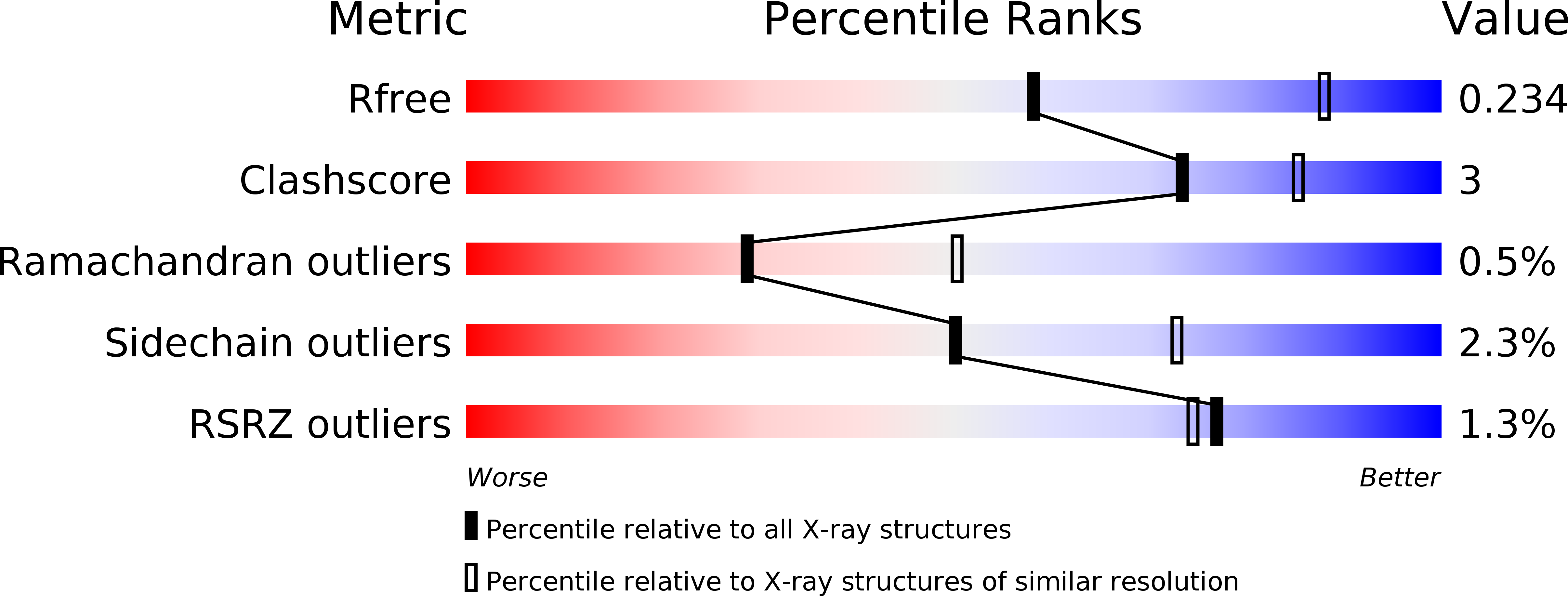
Discovery of a phenylpyrazole amide ROCK inhibitor as a tool molecule for in vivo studies.
Hu, Z., Wang, C., Glunz, P.W., Li, J., Cheadle, N.L., Chen, A.Y., Chen, X.Q., Myers, J.E., Guarino, V.R., Rose, A., Sack, J.S., Sitkoff, D., Taylor, D.S., Xu, S., Yan, C., Zhang, H., Zhang, L., Hennan, J., Adam, L.P., Wexler, R.R., Quan, M.L.(2020) Bioorg Med Chem Lett 30: 127495-127495
- PubMed: 32798651
- DOI: https://doi.org/10.1016/j.bmcl.2020.127495
- Primary Citation of Related Structures:
7JOU, 7JOV - PubMed Abstract:
Structure-activity relationship optimization on a series of phenylpyrazole amides led to the identification of a dual ROCK1 and ROCK2 inhibitor (25) which demonstrated good potency, kinome selectivity and favorable pharmacokinetic profiles. Compound 25 was selected as a tool molecule for in vivo studies including evaluating hemodynamic effects in telemeterized mice, from which moderate decreases in blood pressure were observed.
Organizational Affiliation:
Research & Early Development, Bristol Myers Squibb, P.O. Box 5400, Princeton, NJ 08543-5400, USA. Electronic address: zilun.hu@bms.com.