Inhibitory mechanism of gold analogues against Pseudomonas global virulence factor regulator Vfr
Chew, B.L.A., Luo, D., Yang, L.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| cAMP-activated global transcriptional regulator Vfr | 210 | Pseudomonas aeruginosa PAO1 | Mutation(s): 0 Gene Names: vfr, PA0652 | 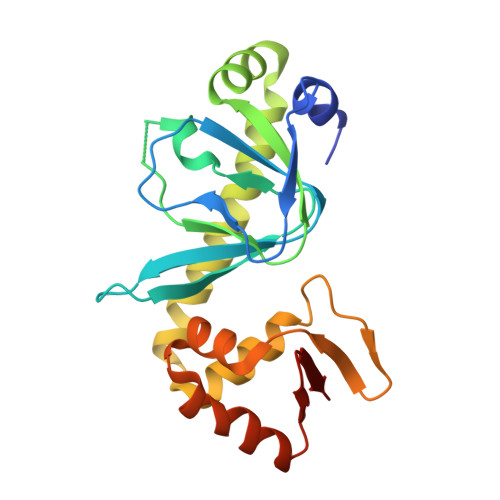 | |
UniProt | |||||
Find proteins for P55222 (Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1)) Explore P55222 Go to UniProtKB: P55222 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P55222 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CMP (Subject of Investigation/LOI) Query on CMP | C [auth A], K [auth B] | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE C10 H12 N5 O6 P IVOMOUWHDPKRLL-KQYNXXCUSA-N |  | ||
| AUF (Subject of Investigation/LOI) Query on AUF | D [auth A], L [auth B] | triethylphosphanuidylgold(1+) C6 H15 Au P NKNLIDCIHVTIMI-UHFFFAOYSA-N |  | ||
| AU (Subject of Investigation/LOI) Query on AU | E [auth A], F [auth A], M [auth B], N [auth B], O [auth B] | GOLD ION Au ZBKIUFWVEIBQRT-UHFFFAOYSA-N |  | ||
| SO4 Query on SO4 | H [auth A], I [auth A], P [auth B], Q [auth B] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| CL (Subject of Investigation/LOI) Query on CL | G [auth A], J [auth B] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 140.264 | α = 90 |
| b = 49.928 | β = 100.145 |
| c = 62.381 | γ = 90 |
| Software Name | Purpose |
|---|---|
| Coot | model building |
| PHENIX | refinement |
| HKL-3000 | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Ministry of Education (MoE, Singapore) | Singapore | 2016-T2-2-097 |