Structural basis for Sarbecovirus ORF6 mediated blockage of nucleocytoplasmic transport
Gao, X., Tian, H., Zhu, K., Li, Q., Hao, W., Wang, L., Qin, B., Deng, H., Cui, S.(2022) Nat Commun 13: 4782
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ORF6 protein | A [auth E], B [auth F] | 63 | Severe acute respiratory syndrome-related coronavirus | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P59634 (Severe acute respiratory syndrome coronavirus) Explore P59634 Go to UniProtKB: P59634 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P59634 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| mRNA export factor | C [auth A], D [auth C] | 368 | Homo sapiens | Mutation(s): 0 Gene Names: RAE1, MRNP41 | 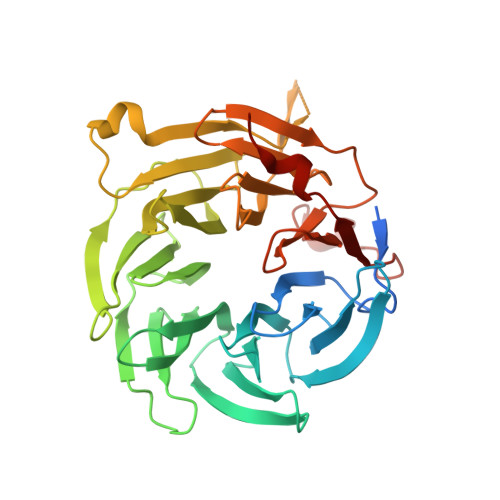 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P78406 (Homo sapiens) Explore P78406 Go to UniProtKB: P78406 | |||||
PHAROS: P78406 GTEx: ENSG00000101146 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P78406 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Nuclear pore complex protein Nup98-Nup96 | E [auth B], F [auth D] | 1,817 | Homo sapiens | Mutation(s): 0 Gene Names: NUP98, ADAR2 EC: 3.4.21 | 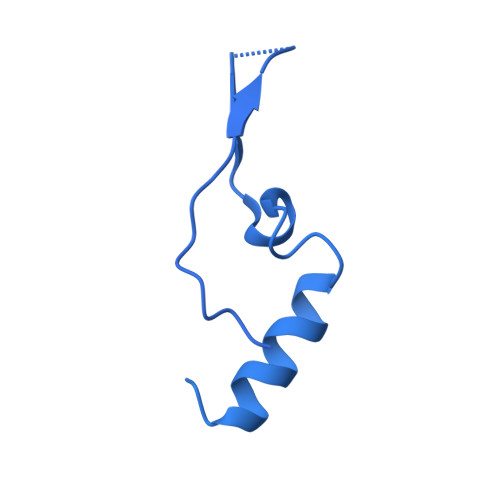 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P52948 (Homo sapiens) Explore P52948 Go to UniProtKB: P52948 | |||||
PHAROS: P52948 GTEx: ENSG00000110713 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P52948 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 189.332 | α = 90 |
| b = 86.794 | β = 92.09 |
| c = 47.905 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| XDS | data scaling |
| Coot | model building |