Tetraphenylporphyrin Enters the Ring: First Example of a Complex between Highly Bulky Porphyrins and a Protein.
Shisaka, Y., Sakakibara, E., Suzuki, K., Stanfield, J.K., Onoda, H., Ueda, G., Hatano, M., Sugimoto, H., Shoji, O.(2022) Chembiochem 23: e202200095-e202200095
- PubMed: 35352458
- DOI: https://doi.org/10.1002/cbic.202200095
- Primary Citation of Related Structures:
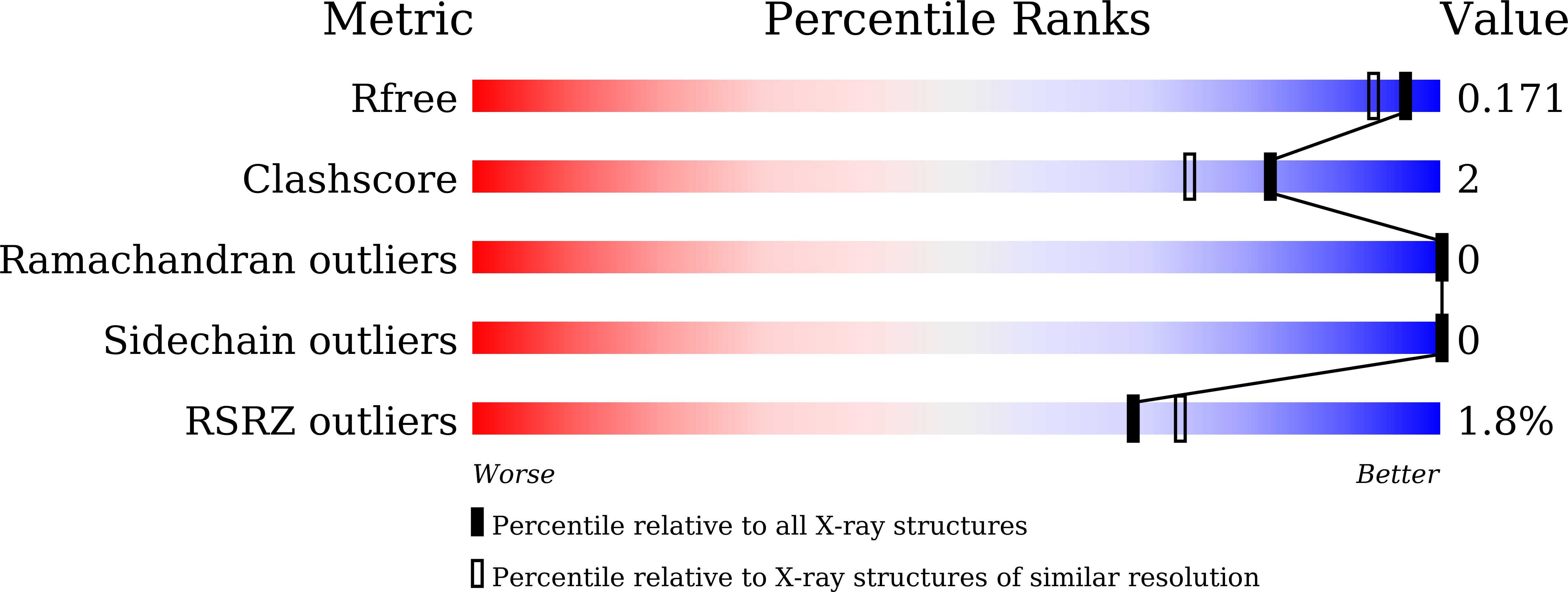
7EMO, 7EMP, 7EMQ, 7EMR, 7EMS, 7EMT, 7EMU, 7EMV, 7EMW, 7VM1 - PubMed Abstract:
Tetraphenylporphyrin (TPP) is a symmetrically substituted synthetic porphyrin whose properties can be readily modified, providing it with significant advantages over naturally occurring porphyrins. Herein, we report the first example of a stable complex between a native biomolecule, the haemoprotein HasA, and TPP as well as its derivatives. The X-ray crystal structures of nine different HasA-TPP complexes were solved at high resolutions. HasA capturing TPP derivatives was also demonstrated to inhibit growth of the opportunistic pathogen Pseudomonas aeruginosa. Mutant variants of HasA binding FeTPP were shown to possess a different mode of coordination, permitting the cyclopropanation of styrene.
Organizational Affiliation:
Department of Chemistry, Graduate School of Science, Nagoya University, Furo-cho, Chikusa-ku, Nagoya, 464-8602, Japan.