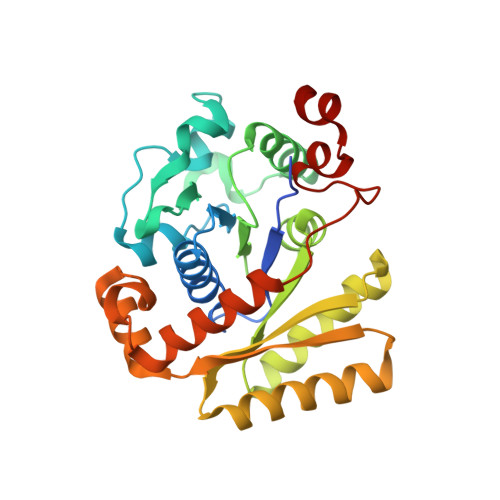
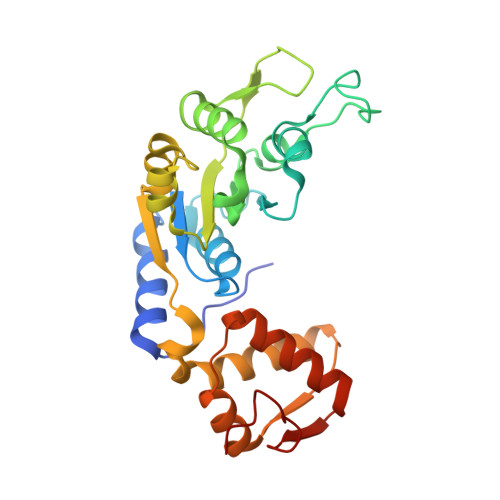
The antiactivator FleN uses an allosteric mechanism to regulate sigma 54 -dependent expression of flagellar genes in Pseudomonas aeruginosa .
Banerjee, P., Raghav, S., Goswami, H.N., Jain, D.(2021) Sci Adv 7: eabj1792-eabj1792
- PubMed: 34669473
- DOI: https://doi.org/10.1126/sciadv.abj1792
- Primary Citation of Related Structures:
7EJW - PubMed Abstract:
Diverse sigma factors associate with the RNA polymerase (RNAP) core enzyme to initiate transcription of specific target genes in bacteria. σ 54 -Mediated transcription uses AAA+ activators that utilize their ATPase activity for transcription initiation. FleQ is a σ 54 -dependent master transcriptional regulator of flagellar genes in Pseudomonas aeruginosa . The ATPase activity of FleQ is regulated via a P-loop ATPase, FleN, through protein-protein interaction. We report a high-resolution crystal structure of the AAA+ domain of FleQ in complex with antiactivator FleN. The data reveal that FleN allosterically prevents ATP binding to FleQ. Furthermore, FleN remodels the region of FleQ essential for engagement with σ 54 for transcription initiation. Disruption of the conserved protein-protein interface, by mutation, shows motility and transcription defects in vivo and multiflagellate phenotype. Our study provides a detailed mechanism used by monoflagellate bacteria to fine-tune the expression of flagellar genes to form and maintain a single flagellum.
Organizational Affiliation:
Transcription Regulation Lab, Regional Centre for Biotechnology, NCR Biotech Science Cluster, 3rd Milestone, Faridabad-Gurgaon Expressway, Faridabad 121001, India.