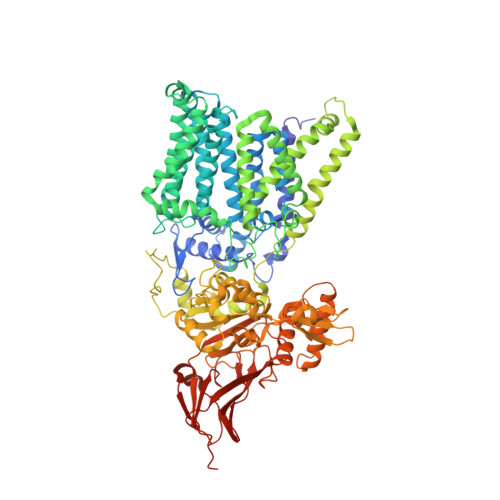
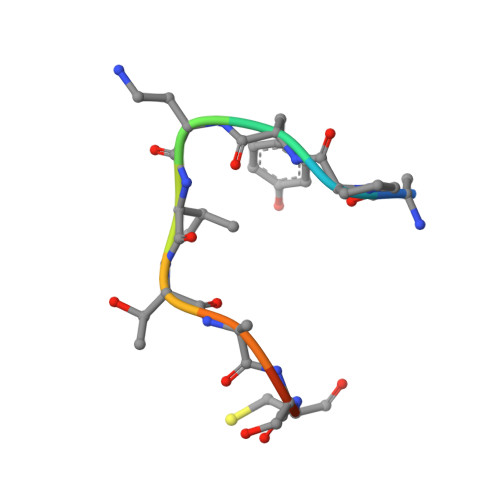
The structure of an archaeal oligosaccharyltransferase provides insight into the strict exclusion of proline from the N-glycosylation sequon.
Taguchi, Y., Yamasaki, T., Ishikawa, M., Kawasaki, Y., Yukimura, R., Mitani, M., Hirata, K., Kohda, D.(2021) Commun Biol 4: 941-941
- PubMed: 34354228
- DOI: https://doi.org/10.1038/s42003-021-02473-8
- Primary Citation of Related Structures:
7E9S - PubMed Abstract:
Oligosaccharyltransferase (OST) catalyzes oligosaccharide transfer to the Asn residue in the N-glycosylation sequon, Asn-X-Ser/Thr, where Pro is strictly excluded at position X. Considering the unique structural properties of proline, this exclusion may not be surprising, but the structural basis for the rejection of Pro residues should be explained explicitly. Here we determined the crystal structure of an archaeal OST in a complex with a sequon-containing peptide and dolichol-phosphate to a 2.7 Å resolution. The sequon part in the peptide forms two inter-chain hydrogen bonds with a conserved amino acid motif, TIXE. We confirmed the essential role of the TIXE motif and the adjacent regions by extensive alanine-scanning of the external loop 5. A Ramachandran plot revealed that the ring structure of the Pro side chain is incompatible with the ϕ backbone dihedral angle around -150° in the rigid sequon-TIXE structure. The present structure clearly provides the structural basis for the exclusion of Pro residues from the N-glycosylation sequon.
Organizational Affiliation:
Division of Structural Biology, Medical Institute of Bioregulation, Kyushu University, Fukuoka, Japan.