An anti-CD98 antibody displaying pH-dependent Fc-mediated tumour-specific activity against multiple cancers in CD98-humanized mice.
Tian, X., Liu, X., Ding, J., Wang, F., Wang, K., Liu, J., Wei, Z., Hao, X., Li, Y., Wei, X., Zhang, H., Sui, J.(2022) Nat Biomed Eng
- PubMed: 36424464
- DOI: https://doi.org/10.1038/s41551-022-00956-5
- Primary Citation of Related Structures:
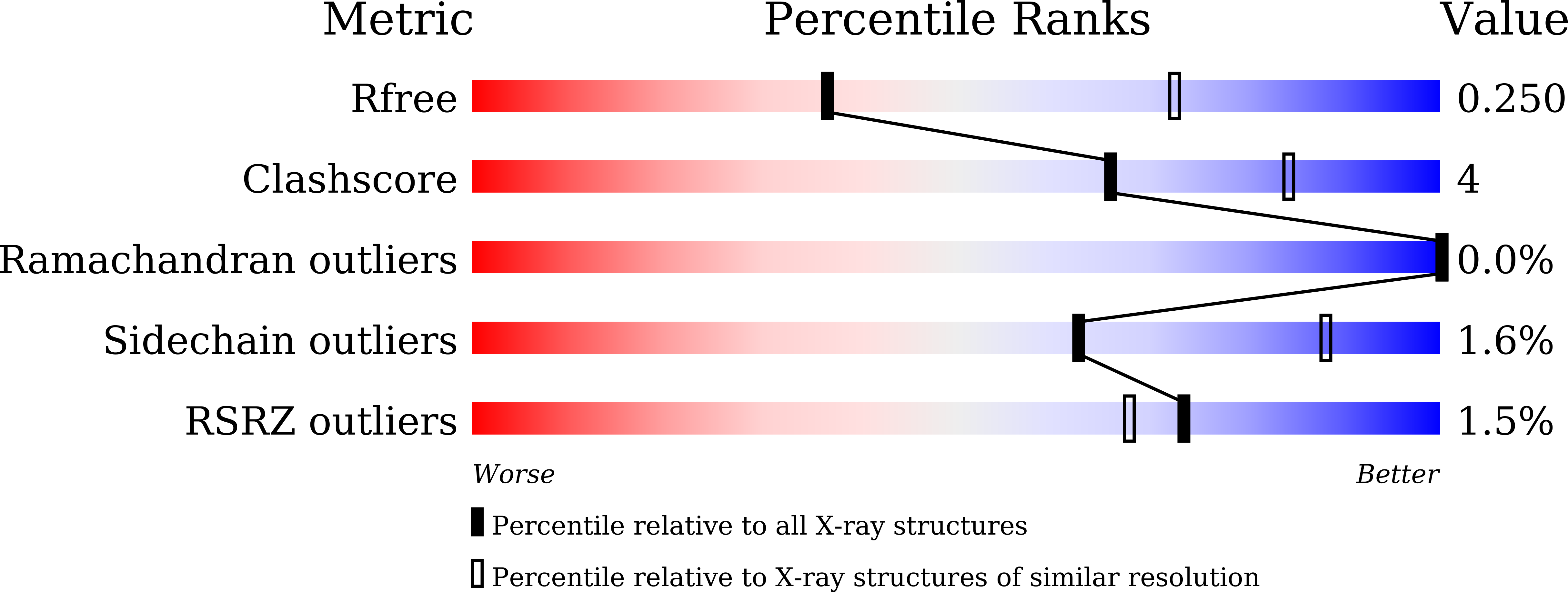
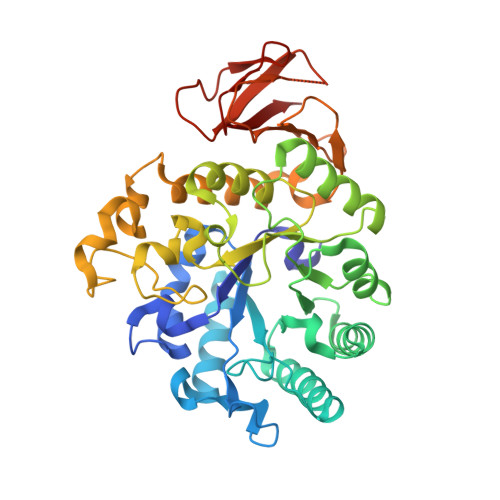
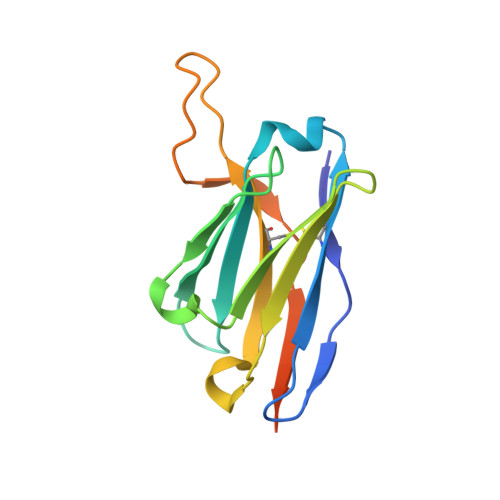
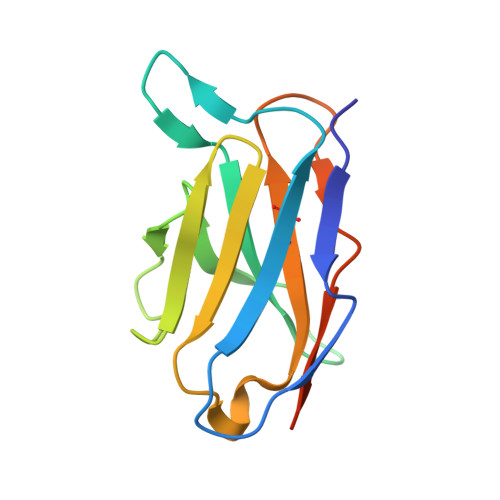
7DF1 - PubMed Abstract:
The cell-surface glycoprotein CD98-a subunit of the LAT1/CD98 amino acid transporter-is an attractive target for cancer immunotherapies, but its widespread expression has hampered the development of CD98-targeting antibody therapeutics. Here we report that an anti-CD98 antibody, identified via the screening of phage-display libraries of CD98 single-chain variable fragments with mutated complementarity-determining regions, preserves the physiological function of CD98 and elicits broad-spectrum crystallizable-fragment (Fc)-mediated anti-tumour activity (requiring Fcγ receptors for immunoglobulins, macrophages, dendritic cells and CD8 + T cells, as well as other components of the innate and adaptive immune systems) in multiple xenograft and syngeneic tumour models established in CD98-humanized mice. We also show that a variant of the anti-CD98 antibody with pH-dependent binding, generated by solving the structure of the antibody-CD98 complex, displayed enhanced tumour-specific activity and pharmacokinetics. pH-dependent antibody variants targeting widely expressed antigens may lead to superior therapeutic outcomes.
Organizational Affiliation:
National Institute of Biological Sciences, Beijing, China.