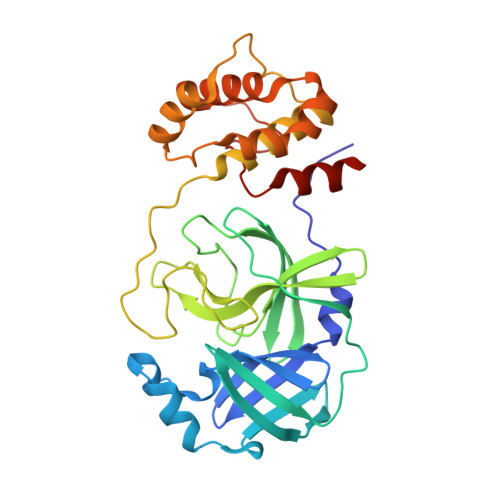
A comparison of Remdesivir versus gold cluster in COVID-19 animal model: A better therapeutic outcome of gold cluster.
He, Z., Ye, F., Zhang, C., Fan, J., Du, Z., Zhao, W., Yuan, Q., Niu, W., Gao, F., He, B., Cao, P., Zhao, L., Gao, X., Gao, X., Sun, B., Dong, Y., Zhao, J., Qi, J., Liang, X.J., Jiang, H., Gong, Y., Tan, W., Gao, X.(2022) Nano Today 44: 101468