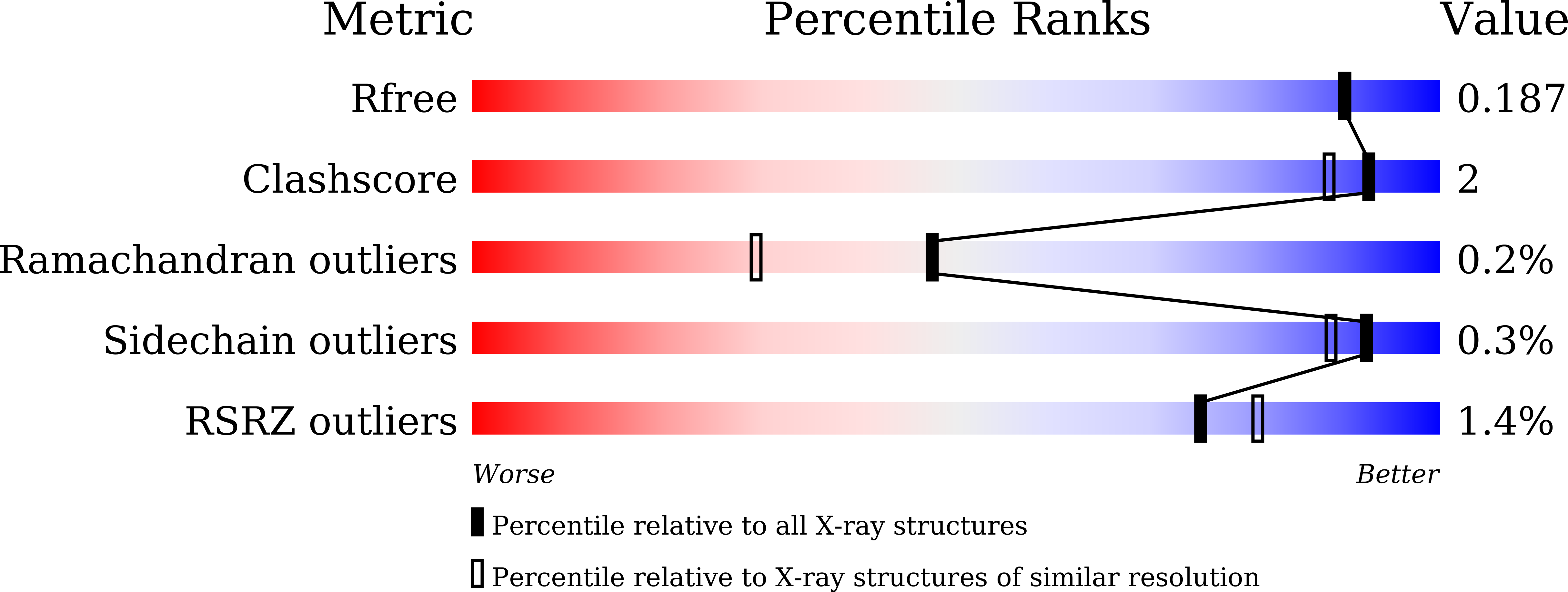
Mutagenesis and structure-based analysis of the role of Tryptophan525 of gamma-glutamyltranspeptidase from Pseudomonas nitroreducens.
Sano, C., Itoh, T., Phumsombat, P., Hayashi, J., Wakayama, M., Hibi, T.(2021) Biochem Biophys Res Commun 534: 286-291
- PubMed: 33288198
- DOI: https://doi.org/10.1016/j.bbrc.2020.11.093
- Primary Citation of Related Structures:
7D9E, 7D9W, 7D9X - PubMed Abstract:
γ-Glutamyltranspeptidase (GGT) is a ubiquitous enzyme that catalyzes the hydrolysis of the γ-glutamyl linkage of γ-glutamyl compounds and the transfer of their γ-glutamyl moiety to acceptor substrates. Pseudomonas nitroreducens GGT (PnGGT) is used for the industrial synthesis of theanine, thus it is important to determine the structural basis of hydrolysis and transfer reactions and identify the acceptor site of PnGGT to improve the efficient of theanine synthesis. Our previous structural studies of PnGGT have revealed that crucial interactions between three amino acid residues, Trp385, Phe417, and Trp525, distinguish PnGGT from other GGTs. Here we report the role of Trp525 in PnGGT based on site-directed mutagenesis and structural analyses. Seven mutant variants of Trp525 were produced (W525F, W525V, W525A, W525G, W525S, W525D, and W525K), with substitution of Trp525 by nonaromatic residues resulting in dramatically reduced hydrolysis activity. All Trp525 mutants exhibited significantly increased transfer activity toward hydroxylamine with hardly any effect on acceptor substrate preference. The crystal structure of PnGGT in complex with the glutamine antagonist, 6-diazo-5-oxo-l-norleucine, revealed that Trp525 is a key residue limiting the movement of water molecules within the PnGGT active site.
Organizational Affiliation:
Department of Biotechnology, College of Life Sciences, Ritsumeikan University, Kusatsu, Shiga, 525-8577, Japan.