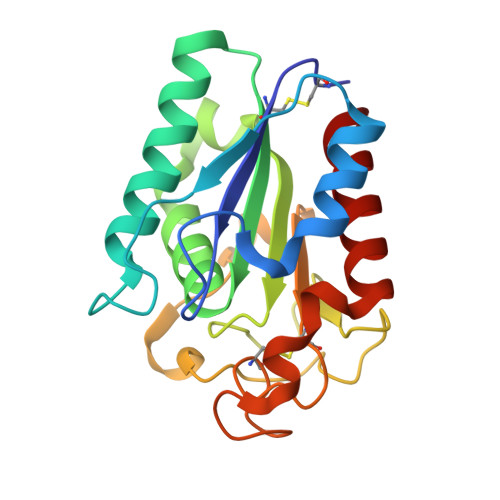
Crystal structure of a biodegradable plastic-degrading cutinase-like enzyme from the phyllosphere yeast, Pseudozyma antarctica, solved by getting the phase from anomalous scattering of uncovalently coordinated arsenic (cacodylate).
Suzuki, K.To be published.