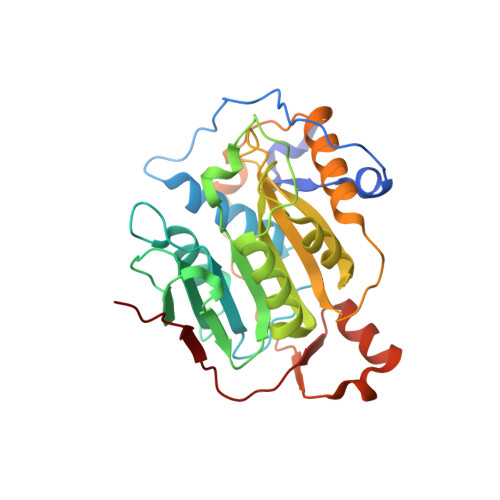
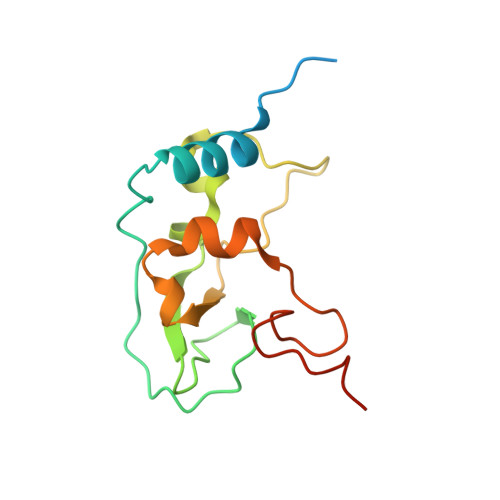
Crystal structure of SARS-CoV-2 nsp10/nsp16 2'-O-methylase and its implication on antiviral drug design.
Lin, S., Chen, H., Ye, F., Chen, Z., Yang, F., Zheng, Y., Cao, Y., Qiao, J., Yang, S., Lu, G.(2020) Signal Transduct Target Ther 5: 131-131
- PubMed: 32728018
- DOI: https://doi.org/10.1038/s41392-020-00241-4
- Primary Citation of Related Structures:
7C2I, 7C2J
Organizational Affiliation:
West China Hospital Emergency Department (WCHED), State Key Laboratory of Biotherapy and Cancer Center, West China Hospital and Collaborative Innovation Center of Biotherapy, Sichuan University, 610041, Chengdu, Sichuan, China.