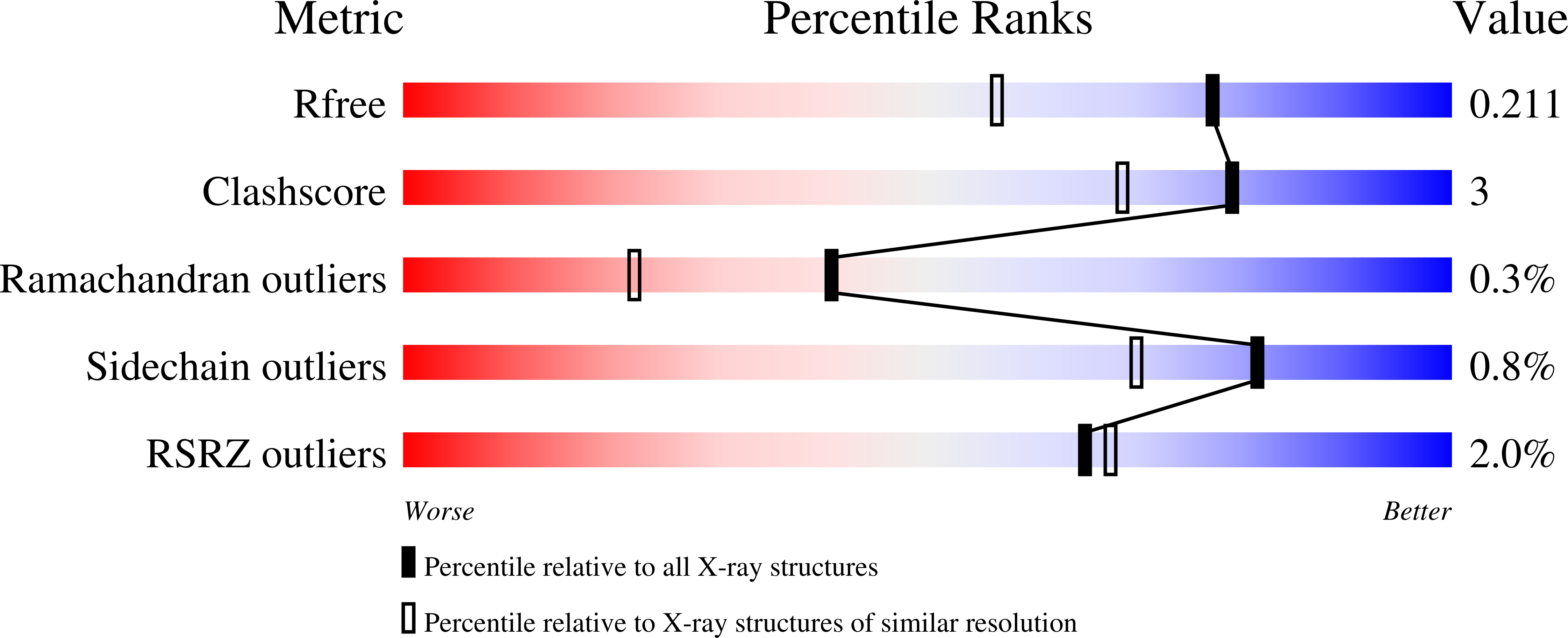
Inhibition mechanism of SARS-CoV-2 main protease by ebselen and its derivatives.
Amporndanai, K., Meng, X., Shang, W., Jin, Z., Rogers, M., Zhao, Y., Rao, Z., Liu, Z.J., Yang, H., Zhang, L., O'Neill, P.M., Samar Hasnain, S.(2021) Nat Commun 12: 3061-3061
- PubMed: 34031399
- DOI: https://doi.org/10.1038/s41467-021-23313-7
- Primary Citation of Related Structures:
7BAJ, 7BAK, 7BAL - PubMed Abstract:
The SARS-CoV-2 pandemic has triggered global efforts to develop therapeutics. The main protease of SARS-CoV-2 (M pro ), critical for viral replication, is a key target for therapeutic development. An organoselenium drug called ebselen has been demonstrated to have potent M pro inhibition and antiviral activity. We have examined the binding modes of ebselen and its derivative in M pro via high resolution co-crystallography and investigated their chemical reactivity via mass spectrometry. Stronger M pro inhibition than ebselen and potent ability to rescue infected cells were observed for a number of derivatives. A free selenium atom bound with cysteine of catalytic dyad has been revealed in crystallographic structures of M pro with ebselen and MR6-31-2 suggesting hydrolysis of the enzyme bound organoselenium covalent adduct and formation of a phenolic by-product, confirmed by mass spectrometry. The target engagement with selenation mechanism of inhibition suggests wider therapeutic applications of these compounds against SARS-CoV-2 and other zoonotic beta-corona viruses.
Organizational Affiliation:
Molecular Biophysics Group, Department of Biochemistry and System Biology, Institute of System, Molecular and Integrative Biology, Faculty of Health and Life Sciences, University of Liverpool, Liverpool, L69 7ZB, UK.