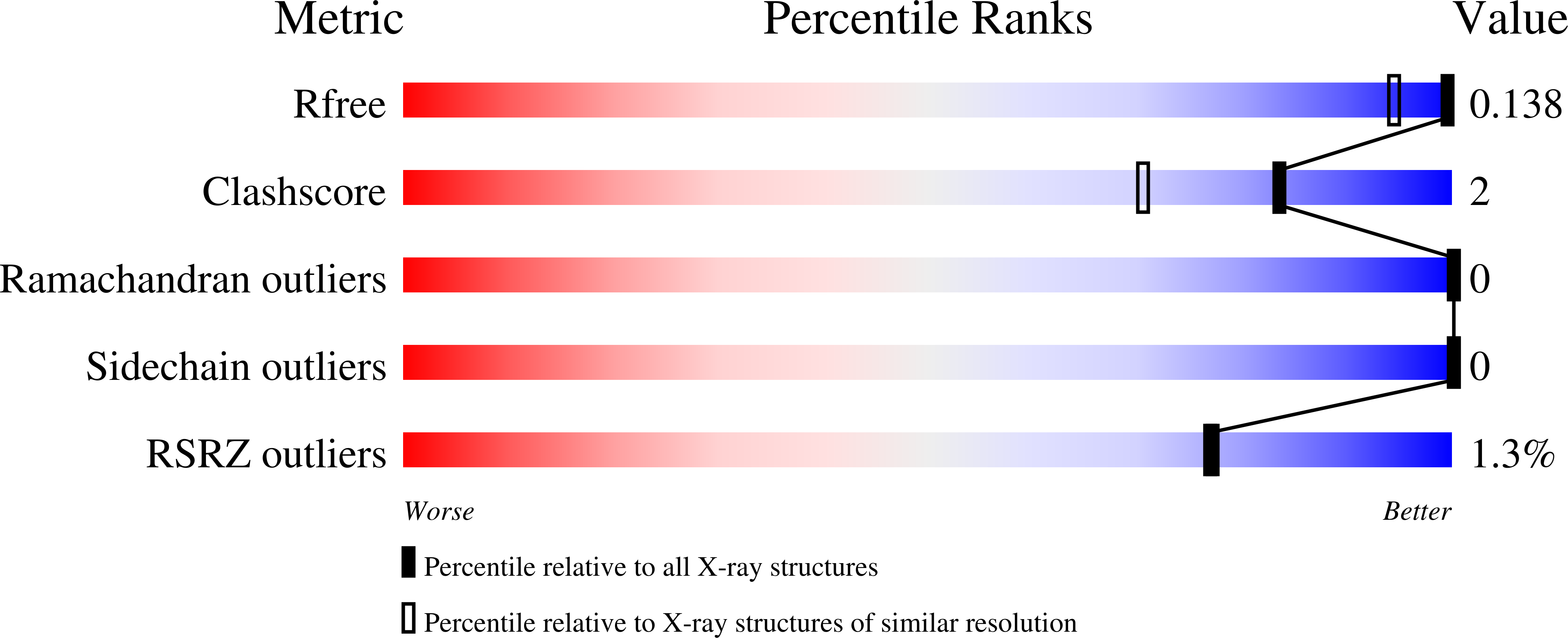
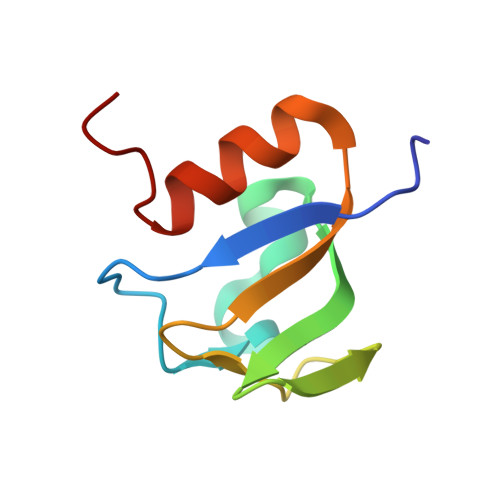
BusR senses bipartite DNA binding motifs by a unique molecular ruler architecture.
Bandera, A.M., Bartho, J., Lammens, K., Drexler, D.J., Kleinschwarzer, J., Hopfner, K.P., Witte, G.(2021) Nucleic Acids Res 49: 10166-10177
- PubMed: 34432045
- DOI: https://doi.org/10.1093/nar/gkab736
- Primary Citation of Related Structures:
7B5T, 7B5U, 7B5W, 7B5Y, 7OZ3 - PubMed Abstract:
The cyclic dinucleotide second messenger c-di-AMP is a major player in regulation of potassium homeostasis and osmolyte transport in a variety of bacteria. Along with various direct interactions with proteins such as potassium channels, the second messenger also specifically binds to transcription factors, thereby altering the processes in the cell on the transcriptional level. We here describe the structural and biochemical characterization of BusR from the human pathogen Streptococcus agalactiae. BusR is a member of a yet structurally uncharacterized subfamily of the GntR family of transcription factors that downregulates transcription of the genes for the BusA (OpuA) glycine-betaine transporter upon c-di-AMP binding. We report crystal structures of full-length BusR, its apo and c-di-AMP bound effector domain, as well as cryo-EM structures of BusR bound to its operator DNA. Our structural data, supported by biochemical and biophysical data, reveal that BusR utilizes a unique domain assembly with a tetrameric coiled-coil in between the binding platforms, serving as a molecular ruler to specifically recognize a 22 bp separated bipartite binding motif. Binding of c-di-AMP to BusR induces a shift in equilibrium from an inactivated towards an activated state that allows BusR to bind the target DNA, leading to transcriptional repression.
Organizational Affiliation:
Gene Center, Ludwig-Maximilians-Universität München, Feodor-Lynen-Str. 25, D-81377 München, Germany.