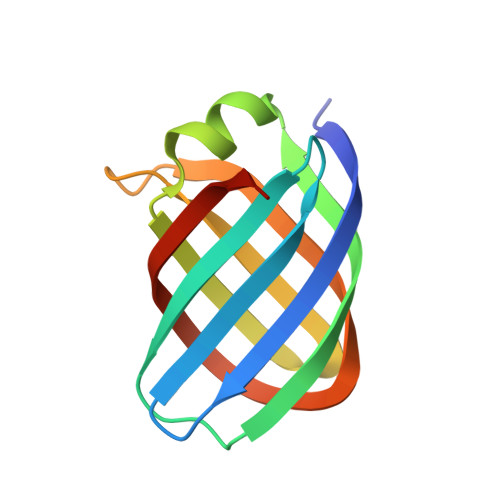
Conformational changes of loops highlight a potential binding site in Rhodococcus equi VapB.
Geerds, C., Haas, A., Niemann, H.H.(2021) Acta Crystallogr F Struct Biol Commun 77: 246-253
- PubMed: 34341190
- DOI: https://doi.org/10.1107/S2053230X2100738X
- Primary Citation of Related Structures:
7B1Z - PubMed Abstract:
Virulence-associated proteins (Vaps) contribute to the virulence of the pathogen Rhodococcus equi, but their mode of action has remained elusive. All Vaps share a conserved core of about 105 amino acids that folds into a compact eight-stranded antiparallel β-barrel with a unique topology. At the top of the barrel, four loops connect the eight β-strands. Previous Vap structures did not show concave surfaces that might serve as a ligand-binding site. Here, the structure of VapB in a new crystal form was determined at 1.71 Å resolution. The asymmetric unit contains two molecules. In one of them, the loop regions at the top of the barrel adopt a different conformation from other Vap structures. An outward movement of the loops results in the formation of a hydrophobic cavity that might act as a ligand-binding site. This lends further support to the hypothesis that the structural similarity between Vaps and avidins suggests a potential binding function for Vaps.
Organizational Affiliation:
Department of Chemistry, Bielefeld University, Universitaetsstrasse 25, 33615 Bielefeld, Germany.