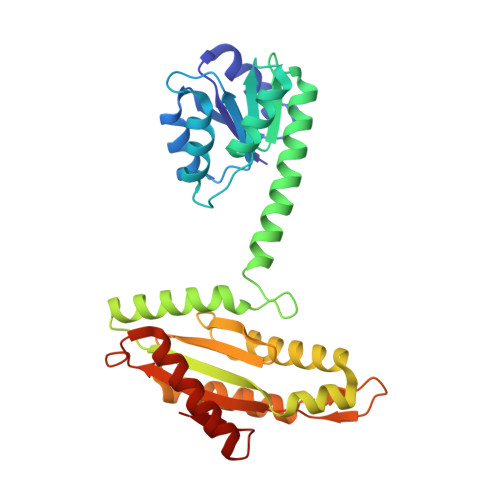
Activation mechanism of a small prototypic Rec-GGDEF diguanylate cyclase.
Teixeira, R.D., Holzschuh, F., Schirmer, T.(2021) Nat Commun 12: 2162-2162
- PubMed: 33846343
- DOI: https://doi.org/10.1038/s41467-021-22492-7
- Primary Citation of Related Structures:
6ZXB, 6ZXC, 6ZXM - PubMed Abstract:
Diguanylate cyclases synthesising the bacterial second messenger c-di-GMP are found to be regulated by a variety of sensory input domains that control the activity of their catalytical GGDEF domain, but how activation proceeds mechanistically is, apart from a few examples, still largely unknown. As part of two-component systems, they are activated by cognate histidine kinases that phosphorylate their Rec input domains. DgcR from Leptospira biflexa is a constitutively dimeric prototype of this class of diguanylate cyclases. Full-length crystal structures reveal that BeF 3 - pseudo-phosphorylation induces a relative rotation of two rigid halves in the Rec domain. This is coupled to a reorganisation of the dimeric structure with concomitant switching of the coiled-coil linker to an alternative heptad register. Finally, the activated register allows the two substrate-loaded GGDEF domains, which are linked to the end of the coiled-coil via a localised hinge, to move into a catalytically competent dimeric arrangement. Bioinformatic analyses suggest that the binary register switch mechanism is utilised by many diguanylate cyclases with N-terminal coiled-coil linkers.
Organizational Affiliation:
Structural Biology, Biozentrum, University of Basel, Basel, Switzerland.