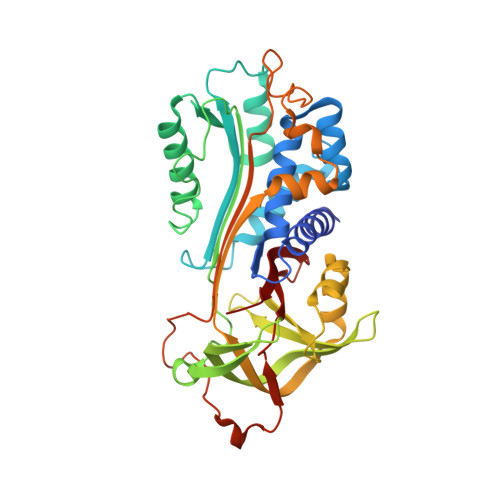
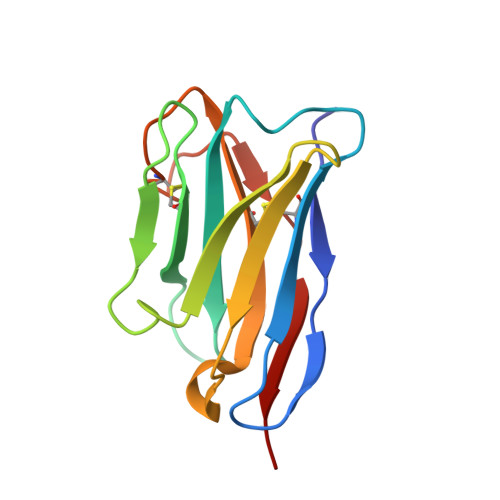
Structural Insights into the Mechanism of a Nanobody That Stabilizes PAI-1 and Modulates Its Activity.
Sillen, M., Weeks, S.D., Strelkov, S.V., Declerck, P.J.(2020) Int J Mol Sci 21
- PubMed: 32824134
- DOI: https://doi.org/10.3390/ijms21165859
- Primary Citation of Related Structures:
6ZRV - PubMed Abstract:
Plasminogen activator inhibitor-1 (PAI-1) is the main physiological inhibitor of tissue-type (tPA) and urokinase-type (uPA) plasminogen activators (PAs). Apart from being critically involved in fibrinolysis and wound healing, emerging evidence indicates that PAI-1 plays an important role in many diseases, including cardiovascular disease, tissue fibrosis, and cancer. Targeting PAI-1 is therefore a promising therapeutic strategy in PAI-1 related pathologies. Despite ongoing efforts no PAI-1 inhibitors were approved to date for therapeutic use in humans. A better understanding of the molecular mechanisms of PAI-1 inhibition is therefore necessary to guide the rational design of PAI-1 modulators. Here, we present a 1.9 Å crystal structure of PAI-1 in complex with an inhibitory nanobody VHH-s-a93 (Nb93). Structural analysis in combination with biochemical characterization reveals that Nb93 directly interferes with PAI-1/PA complex formation and stabilizes the active conformation of the PAI-1 molecule.
Organizational Affiliation:
Laboratory for Therapeutic and Diagnostic Antibodies, Department of Pharmaceutical and Pharmacological Sciences, KU Leuven, B-3000 Leuven, Belgium.