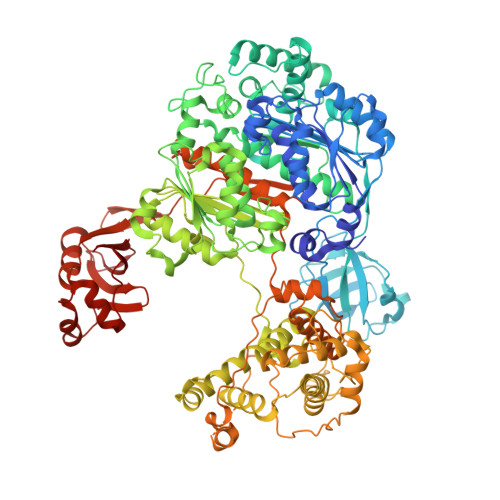
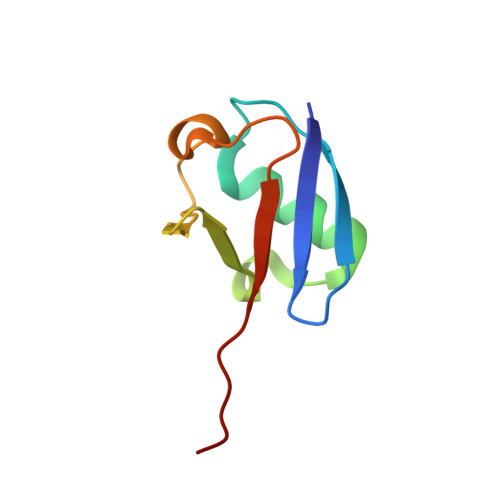
Development of ADPribosyl Ubiquitin Analogues to Study Enzymes Involved in Legionella Infection.
Kim, R.Q., Misra, M., Gonzalez, A., Tomaskovic, I., Shin, D., Schindelin, H., Filippov, D.V., Ovaa, H., Dikic, I., van der Heden van Noort, G.J.(2021) Chemistry 27: 2506-2512
- PubMed: 33075184
- DOI: https://doi.org/10.1002/chem.202004590
- Primary Citation of Related Structures:
6ZQH - PubMed Abstract:
Legionnaires' disease is caused by infection with the intracellularly replicating Gram-negative bacterium Legionella pneumophila. This pathogen uses an unconventional way of ubiquitinating host proteins by generating a phosphoribosyl linkage between substrate proteins and ubiquitin by making use of an ADPribosylated ubiquitin (Ub ADPr ) intermediate. The family of SidE effector enzymes that catalyze this reaction is counteracted by Legionella hydrolases, which are called Dups. This unusual ubiquitination process is important for Legionella proliferation and understanding these processes on a molecular level might prove invaluable in finding new treatments. Herein, a modular approach is used for the synthesis of triazole-linked Ub ADPr , and analogues thereof, and their affinity towards the hydrolase DupA is determined and hydrolysis rates are compared to natively linked Ub ADPr . The inhibitory effects of modified Ub on the canonical eukaryotic E1-enzyme Uba1 are investigated and rationalized in the context of a high-resolution crystal structure reported herein. Finally, it is shown that synthetic Ub ADPr analogues can be used to effectively pull-down overexpressed DupA from cell lysate.
Organizational Affiliation:
Oncode Institute and Department of Cell and Chemical Biology, Leiden University Medical Centre, Einthovenweg 20, 2333 ZC, Leiden, The Netherlands.