Intermolecular latency regulates the essential C-terminal signal peptidase and sortase of the Porphyromonas gingivalis type-IX secretion system.
Mizgalska, D., Goulas, T., Rodriguez-Banqueri, A., Veillard, F., Madej, M., Malecka, E., Szczesniak, K., Ksiazek, M., Widziolek, M., Guevara, T., Eckhard, U., Sola, M., Potempa, J., Gomis-Ruth, F.X.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 34593635
- DOI: https://doi.org/10.1073/pnas.2103573118
- Primary Citation of Related Structures:
6ZA2 - PubMed Abstract:
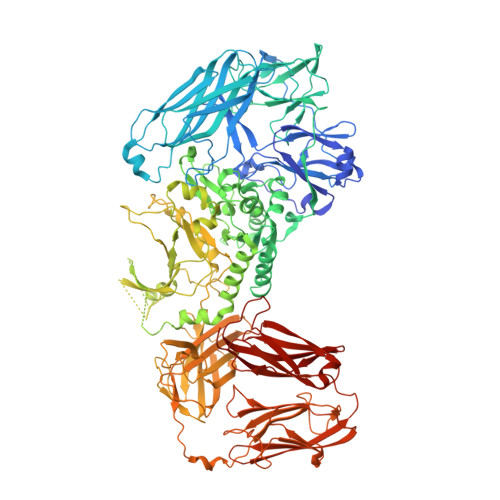
Porphyromonas gingivalis is a keystone pathogen of the human dysbiotic oral microbiome that causes severe periodontitis. It employs a type-IX secretion system (T9SS) to shuttle proteins across the outer membrane (OM) for virulence. Uniquely, T9SS cargoes carry a C-terminal domain (CTD) as a secretion signal, which is cleaved and replaced with anionic lipopolysaccharide by transpeptidation for extracellular anchorage to the OM. Both reactions are carried out by PorU, the only known dual-function, C-terminal signal peptidase and sortase. PorU is itself secreted by the T9SS, but its CTD is not removed; instead, intact PorU combines with PorQ, PorV, and PorZ in the OM-inserted "attachment complex." Herein, we revealed that PorU transits between active monomers and latent dimers and solved the crystal structure of the ∼260-kDa dimer. PorU has an elongated shape ∼130 Å in length and consists of seven domains. The first three form an intertwined N-terminal cluster likely engaged in substrate binding. They are followed by a gingipain-type catalytic domain (CD), two immunoglobulin-like domains (IGL), and the CTD. In the first IGL, a long "latency β-hairpin" protrudes ∼30 Å from the surface to form an intermolecular β-barrel with β-strands from the symmetric CD, which is in a latent conformation. Homology modeling of the competent CD followed by in vivo validation through a cohort of mutant strains revealed that PorU is transported and functions as a monomer through a C 690 /H 657 catalytic dyad. Thus, dimerization is an intermolecular mechanism for PorU regulation to prevent untimely activity until joining the attachment complex.
Organizational Affiliation:
Department of Microbiology, Faculty of Biochemistry, Biophysics and Biotechnology, Jagiellonian University, 30-387 Kraków, Poland.