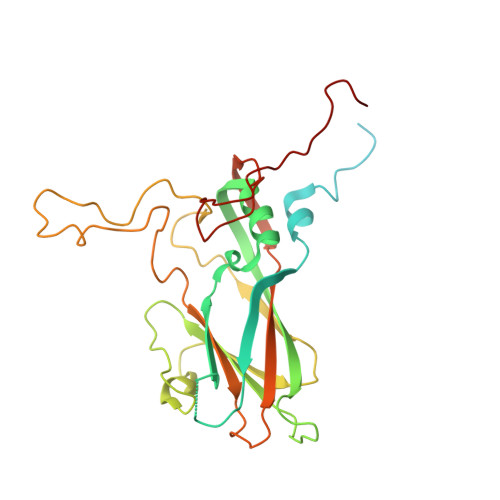
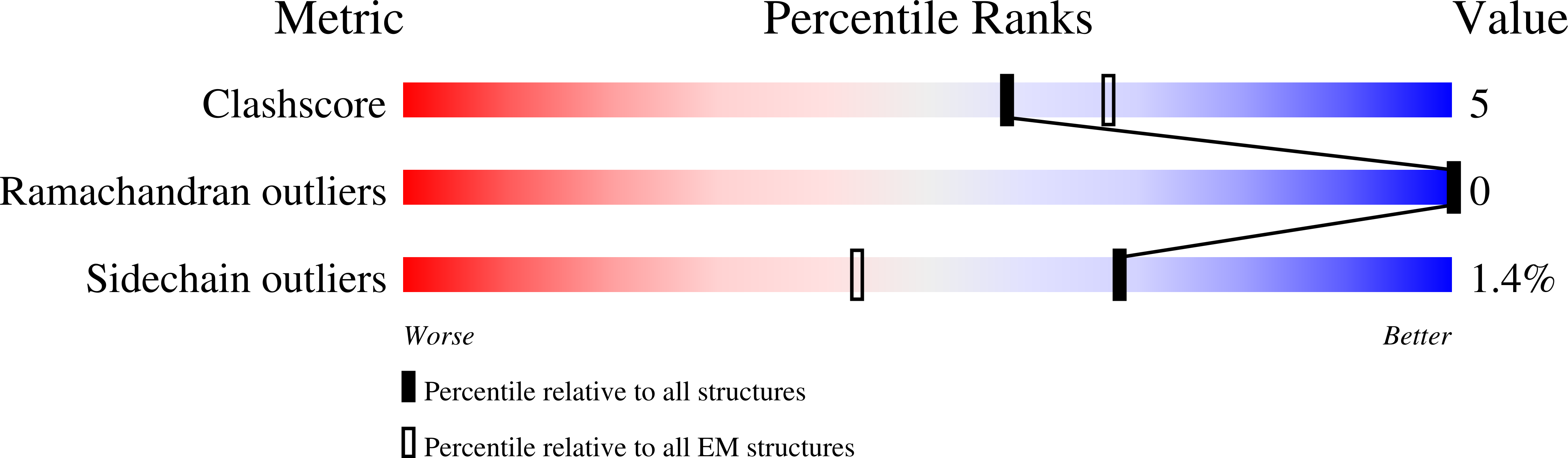Mammalian expression of virus-like particles as a proof of principle for next generation polio vaccines.
Bahar, M.W., Porta, C., Fox, H., Macadam, A.J., Fry, E.E., Stuart, D.I.(2021) NPJ Vaccines 6: 5-5
- PubMed: 33420068
- DOI: https://doi.org/10.1038/s41541-020-00267-3
- Primary Citation of Related Structures:
6Z6W - PubMed Abstract:
Global vaccination programs using live-attenuated oral and inactivated polio vaccine (OPV and IPV) have almost eradicated poliovirus (PV) but these vaccines or their production pose significant risk in a polio-free world. Recombinant PV virus-like particles (VLPs), lacking the viral genome, represent safe next-generation vaccines, however their production requires optimisation. Here we present an efficient mammalian expression strategy producing good yields of wild-type PV VLPs for all three serotypes and a thermostabilised variant for PV3. Whilst the wild-type VLPs were predominantly in the non-native C-antigenic form, the thermostabilised PV3 VLPs adopted the native D-antigenic conformation eliciting neutralising antibody titres equivalent to the current IPV and were indistinguishable from natural empty particles by cryo-electron microscopy with a similar stabilising lipidic pocket-factor in the VP1 β-barrel. This factor may not be available in alternative expression systems, which may require synthetic pocket-binding factors. VLPs equivalent to these mammalian expressed thermostabilized particles, represent safer non-infectious vaccine candidates for the post-eradication era.
Organizational Affiliation:
Division of Structural Biology, University of Oxford, The Henry Wellcome Building for Genomic Medicine, Headington, Oxford, OX3 7BN, UK. mwbahar@strubi.ox.ac.uk.