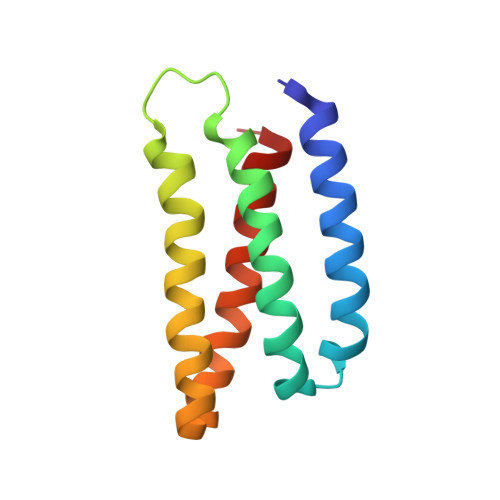
Design of buried charged networks in artificial proteins.
Baumgart, M., Ropke, M., Muhlbauer, M.E., Asami, S., Mader, S.L., Fredriksson, K., Groll, M., Gamiz-Hernandez, A.P., Kaila, V.R.I.(2021) Nat Commun 12: 1895-1895
- PubMed: 33767131
- DOI: https://doi.org/10.1038/s41467-021-21909-7
- Primary Citation of Related Structures:
6Z35 - PubMed Abstract:
Soluble proteins are universally packed with a hydrophobic core and a polar surface that drive the protein folding process. Yet charged networks within the central protein core are often indispensable for the biological function. Here, we show that natural buried ion-pairs are stabilised by amphiphilic residues that electrostatically shield the charged motif from its surroundings to gain structural stability. To explore this effect, we build artificial proteins with buried ion-pairs by combining directed computational design and biophysical experiments. Our findings illustrate how perturbation in charged networks can introduce structural rearrangements to compensate for desolvation effects. We validate the physical principles by resolving high-resolution atomic structures of the artificial proteins that are resistant towards unfolding at extreme temperatures and harsh chemical conditions. Our findings provide a molecular understanding of functional charged networks and how point mutations may alter the protein's conformational landscape.
Organizational Affiliation:
Center for Integrated Protein Science Munich (CIPSM) at the Department Chemie, Technische Universität München, Lichtenbergstraße 4, 85748, Garching, Germany.