High-resolution cryo-EM structure of urease from the pathogen Yersinia enterocolitica.
Righetto, R.D., Anton, L., Adaixo, R., Jakob, R.P., Zivanov, J., Mahi, M.A., Ringler, P., Schwede, T., Maier, T., Stahlberg, H.(2020) Nat Commun 11: 5101-5101
- PubMed: 33037208
- DOI: https://doi.org/10.1038/s41467-020-18870-2
- Primary Citation of Related Structures:
6YL3 - PubMed Abstract:
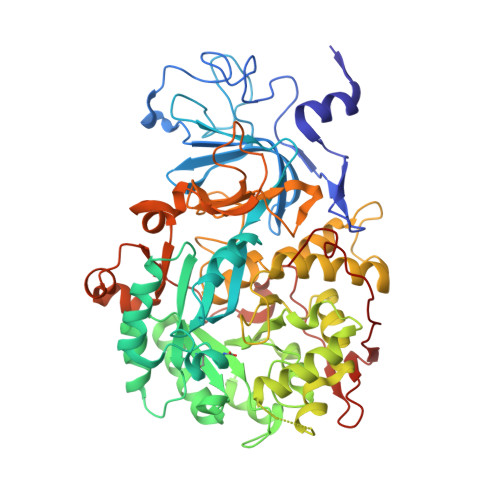
Urease converts urea into ammonia and carbon dioxide and makes urea available as a nitrogen source for all forms of life except animals. In human bacterial pathogens, ureases also aid in the invasion of acidic environments such as the stomach by raising the surrounding pH. Here, we report the structure of urease from the pathogen Yersinia enterocolitica at 2 Å resolution from cryo-electron microscopy. Y. enterocolitica urease is a dodecameric assembly of a trimer of three protein chains, ureA, ureB and ureC. The high data quality enables detailed visualization of the urease bimetal active site and of the impact of radiation damage. The obtained structure is of sufficient quality to support drug development efforts.
Organizational Affiliation:
Center for Cellular Imaging and NanoAnalytics, Biozentrum, University of Basel, Mattenstrasse 26, CH-4058, Basel, Switzerland.