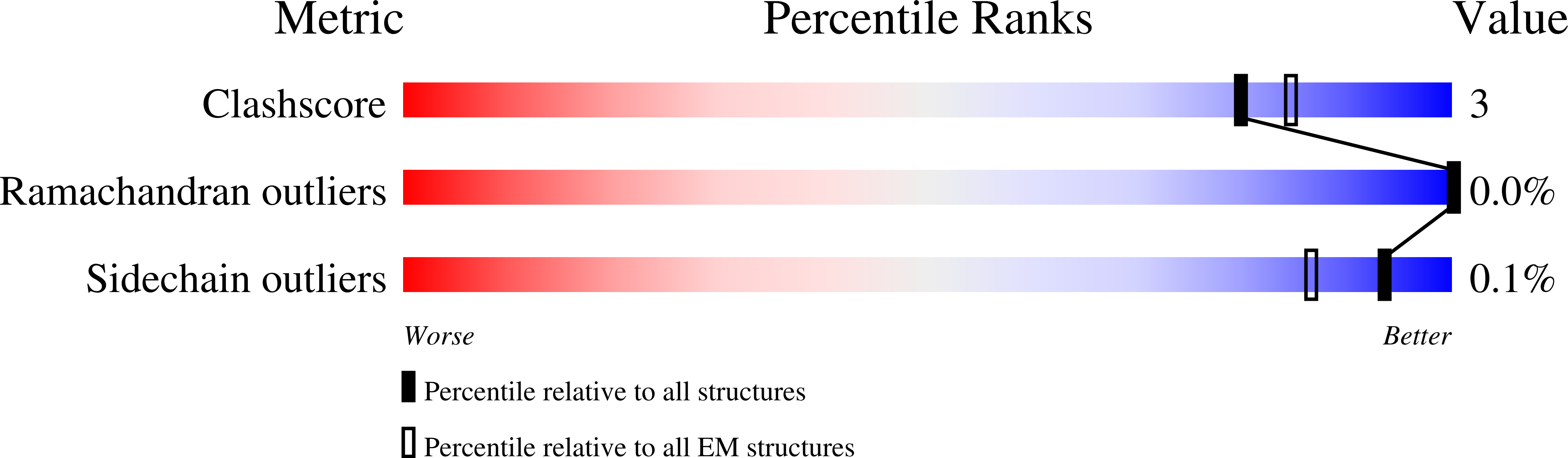Mitochondrial complex I structure reveals ordered water molecules for catalysis and proton translocation.
Grba, D.N., Hirst, J.(2020) Nat Struct Mol Biol 27: 892-900
- PubMed: 32747785
- DOI: https://doi.org/10.1038/s41594-020-0473-x
- Primary Citation of Related Structures:
6YJ4 - PubMed Abstract:
Mitochondrial complex I powers ATP synthesis by oxidative phosphorylation, exploiting the energy from ubiquinone reduction by NADH to drive protons across the energy-transducing inner membrane. Recent cryo-EM analyses of mammalian and yeast complex I have revolutionized structural and mechanistic knowledge and defined structures in different functional states. Here, we describe a 2.7-Å-resolution structure of the 42-subunit complex I from the yeast Yarrowia lipolytica containing 275 structured water molecules. We identify a proton-relay pathway for ubiquinone reduction and water molecules that connect mechanistically crucial elements and constitute proton-translocation pathways through the membrane. By comparison with known structures, we deconvolute structural changes governing the mammalian 'deactive transition' (relevant to ischemia-reperfusion injury) and their effects on the ubiquinone-binding site and a connected cavity in ND1. Our structure thus provides important insights into catalysis by this enigmatic respiratory machine.
Organizational Affiliation:
The Medical Research Council Mitochondrial Biology Unit, University of Cambridge, Cambridge, UK.