Structure-based evolution of a promiscuous inhibitor to a selective stabilizer of protein-protein interactions.
Sijbesma, E., Visser, E., Plitzko, K., Thiel, P., Milroy, L.G., Kaiser, M., Brunsveld, L., Ottmann, C.(2020) Nat Commun 11: 3954-3954
- PubMed: 32770072
- DOI: https://doi.org/10.1038/s41467-020-17741-0
- Primary Citation of Related Structures:
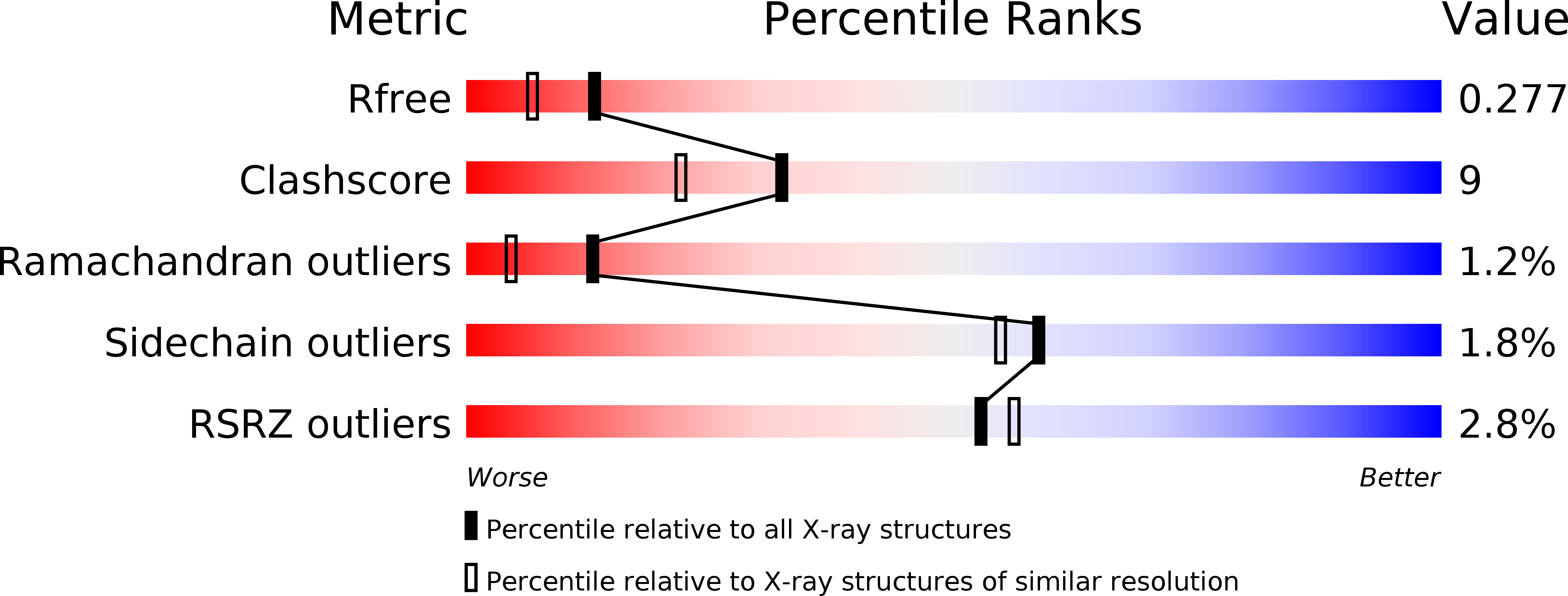
6YGJ - PubMed Abstract:
The systematic stabilization of protein-protein interactions (PPI) has great potential as innovative drug discovery strategy to target novel and hard-to-drug protein classes. The current lack of chemical starting points and focused screening opportunities limits the identification of small molecule stabilizers that engage two proteins simultaneously. Starting from our previously described virtual screening strategy to identify inhibitors of 14-3-3 proteins, we report a conceptual molecular docking approach providing concrete entries for discovery and rational optimization of stabilizers for the interaction of 14-3-3 with the carbohydrate-response element-binding protein (ChREBP). X-ray crystallography reveals a distinct difference in the binding modes between weak and general inhibitors of 14-3-3 complexes and a specific, potent stabilizer of the 14-3-3/ChREBP complex. Structure-guided stabilizer optimization results in selective, up to 26-fold enhancement of the 14-3-3/ChREBP interaction. This study demonstrates the potential of rational design approaches for the development of selective PPI stabilizers starting from weak, promiscuous PPI inhibitors.
Organizational Affiliation:
Laboratory of Chemical Biology and Institute for Complex Molecular Systems (ICMS), Department of Biomedical Engineering, Eindhoven University of Technology, Eindhoven, Netherlands.