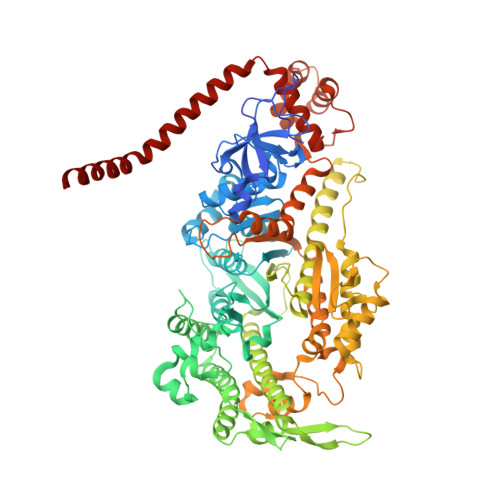
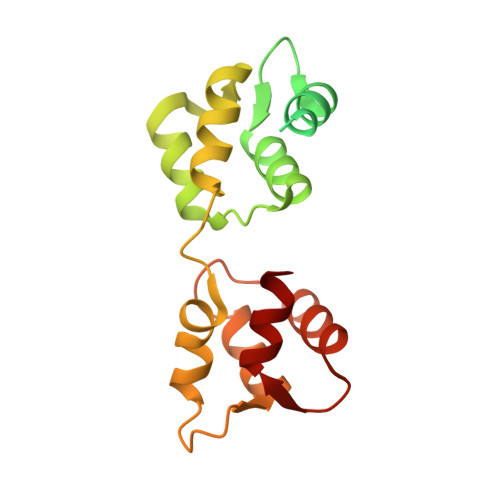
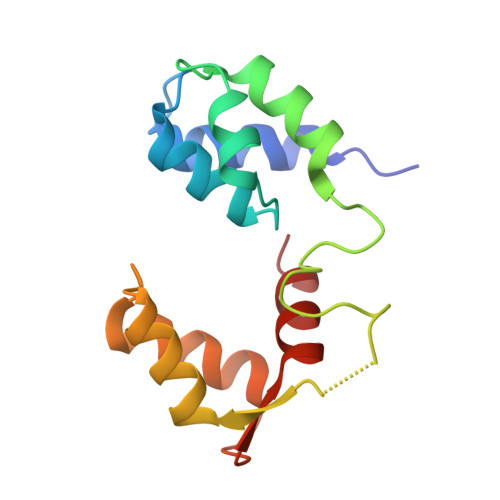
Full-length Plasmodium falciparum myosin A and essential light chain PfELC structures provide new anti-malarial targets.
Moussaoui, D., Robblee, J.P., Auguin, D., Krementsova, E.B., Haase, S., Blake, T.C.A., Baum, J., Robert-Paganin, J., Trybus, K.M., Houdusse, A.(2020) Elife 9
- PubMed: 33046215
- DOI: https://doi.org/10.7554/eLife.60581
- Primary Citation of Related Structures:
6YCX, 6YCY, 6YCZ - PubMed Abstract:
Parasites from the genus Plasmodium are the causative agents of malaria. The mobility, infectivity, and ultimately pathogenesis of Plasmodium falciparum rely on a macromolecular complex, called the glideosome. At the core of the glideosome is an essential and divergent Myosin A motor (PfMyoA), a first order drug target against malaria. Here, we present the full-length structure of PfMyoA in two states of its motor cycle. We report novel interactions that are essential for motor priming and the mode of recognition of its two light chains (PfELC and MTIP) by two degenerate IQ motifs. Kinetic and motility assays using PfMyoA variants, along with molecular dynamics, demonstrate how specific priming and atypical sequence adaptations tune the motor's mechano-chemical properties. Supported by evidence for an essential role of the PfELC in malaria pathogenesis, these structures provide a blueprint for the design of future anti-malarials targeting both the glideosome motor and its regulatory elements.
Organizational Affiliation:
Structural Motility, Institut Curie, Paris Université Sciences et Lettres, Sorbonne Université, CNRS UMR144, Paris, France.