Diphenylurea-derived cytokinin oxidase/dehydrogenase inhibitors for biotechnology and agriculture.
Nisler, J., Kopecny, D., Pekna, Z., Koncitikova, R., Koprna, R., Murvanidze, N., Werbrouck, S.P.O., Havlicek, L., De Diego, N., Kopecna, M., Wimmer, Z., Briozzo, P., Morera, S., Zalabak, D., Spichal, L., Strnad, M.(2021) J Exp Bot 72: 355-370
- PubMed: 32945834
- DOI: https://doi.org/10.1093/jxb/eraa437
- Primary Citation of Related Structures:
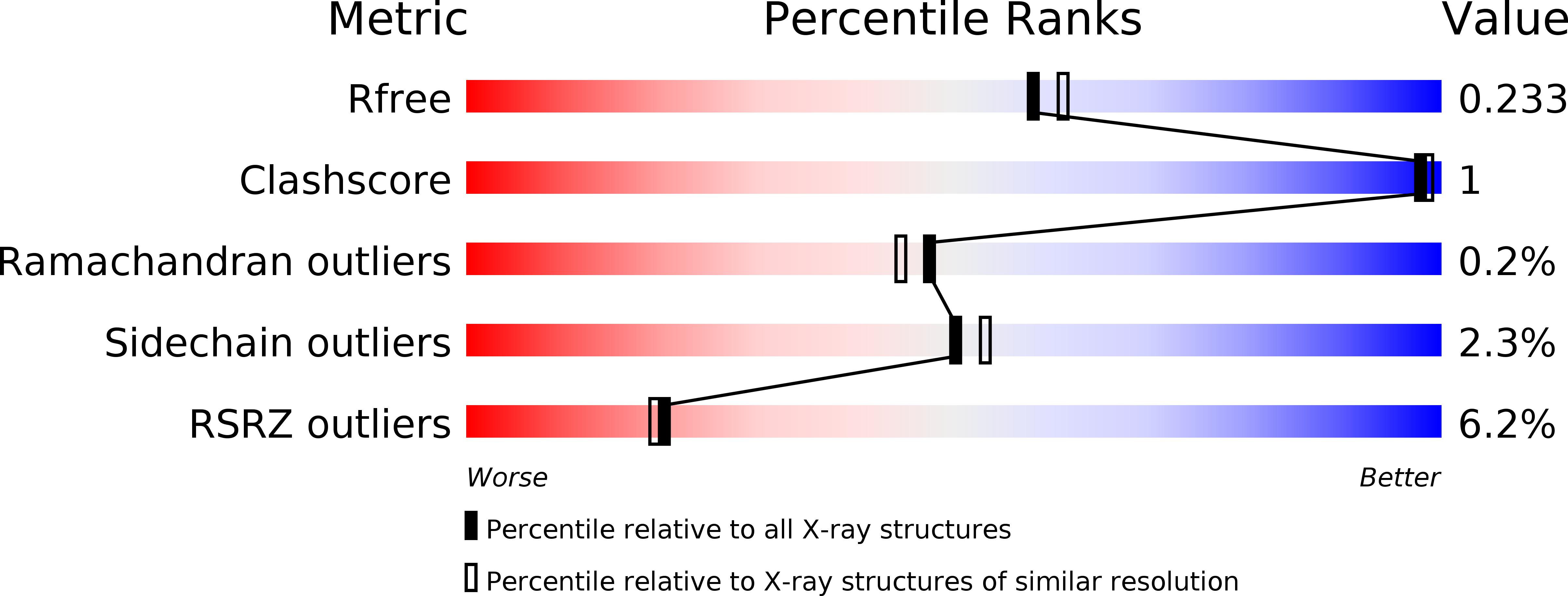
6YAO, 6YAP, 6YAQ - PubMed Abstract:
Increasing crop productivity is our major challenge if we are to meet global needs for food, fodder and fuel. Controlling the content of the plant hormone cytokinin is a method of improving plant productivity. Cytokinin oxidase/dehydrogenase (CKO/CKX) is a major target in this regard because it degrades cytokinins. Here, we describe the synthesis and biological activities of new CKX inhibitors derived mainly from diphenylurea. They were tested on four CKX isoforms from maize and Arabidopsis, where the best compounds showed IC50 values in the 10-8 M concentration range. The binding mode of the most efficient inhibitors was characterized from high-resolution crystal complexed structures. Although these compounds do not possess intrinsic cytokinin activity, we have demonstrated their tremendous potential for use in the plant tissue culture industry as well as in agriculture. We have identified a key substance, compound 19, which not only increases stress resistance and seed yield in Arabidopsis, but also improves the yield of wheat, barley and rapeseed grains under field conditions. Our findings reveal that modulation of cytokinin levels via CKX inhibition can positively affect plant growth, development and yield, and prove that CKX inhibitors can be an attractive target in plant biotechnology and agriculture.
Organizational Affiliation:
Laboratory of Growth Regulators, Institute of Experimental Botany of the Czech Academy of Sciences & Palacký University, Šlechtitelů 27, Olomouc, Czech Republic.