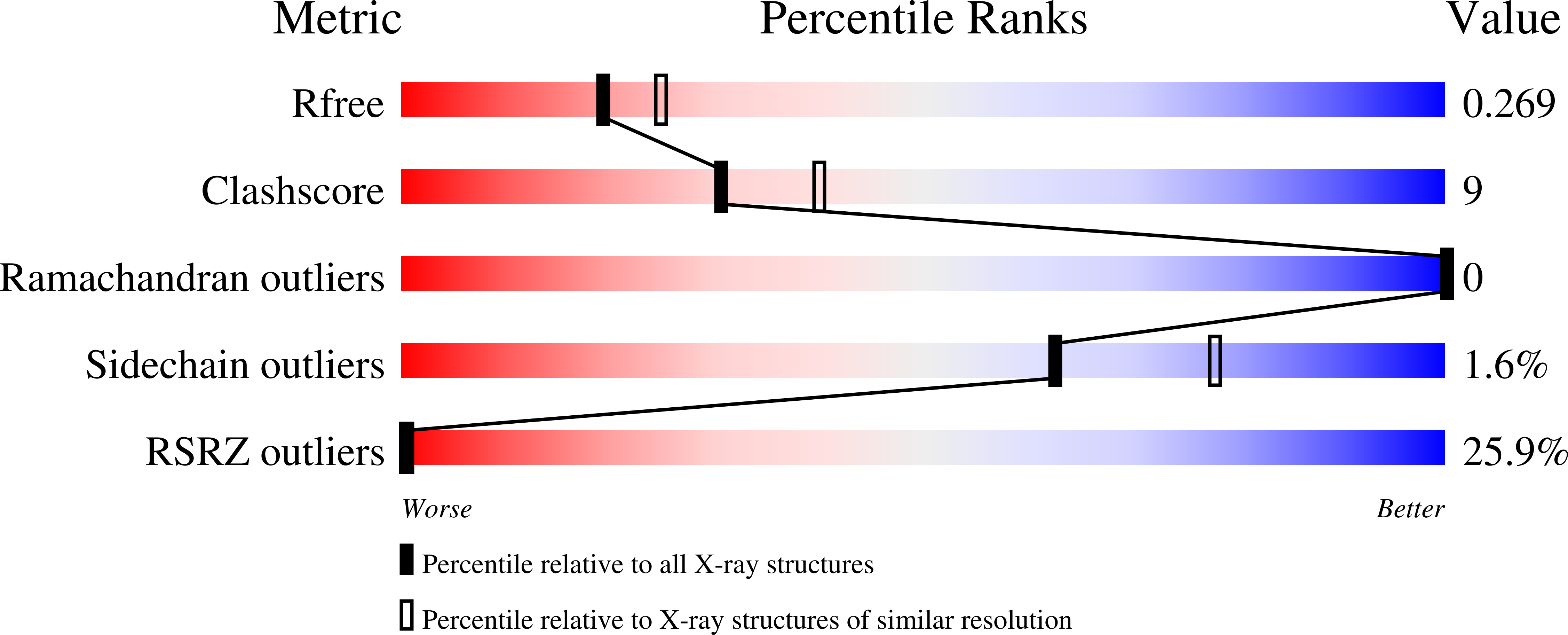
Kinetic and Structural Characterization of the Self-Labeling Protein Tags HaloTag7, SNAP-tag, and CLIP-tag.
Wilhelm, J., Kuhn, S., Tarnawski, M., Gotthard, G., Tunnermann, J., Tanzer, T., Karpenko, J., Mertes, N., Xue, L., Uhrig, U., Reinstein, J., Hiblot, J., Johnsson, K.(2021) Biochemistry 60: 2560-2575
- PubMed: 34339177
- DOI: https://doi.org/10.1021/acs.biochem.1c00258
- Primary Citation of Related Structures:
6Y7A, 6Y7B, 6Y8P, 6ZCC - PubMed Abstract:
The self-labeling protein tags (SLPs) HaloTag7, SNAP-tag, and CLIP-tag allow the covalent labeling of fusion proteins with synthetic molecules for applications in bioimaging and biotechnology. To guide the selection of an SLP-substrate pair and provide guidelines for the design of substrates, we report a systematic and comparative study of the labeling kinetics and substrate specificities of HaloTag7, SNAP-tag, and CLIP-tag. HaloTag7 reaches almost diffusion-limited labeling rate constants with certain rhodamine substrates, which are more than 2 orders of magnitude higher than those of SNAP-tag for the corresponding substrates. SNAP-tag labeling rate constants, however, are less affected by the structure of the label than those of HaloTag7, which vary over 6 orders of magnitude for commonly employed substrates. Determining the crystal structures of HaloTag7 and SNAP-tag labeled with fluorescent substrates allowed us to rationalize their substrate preferences. We also demonstrate how these insights can be exploited to design substrates with improved labeling kinetics.
Organizational Affiliation:
Department of Chemical Biology, Max Planck Institute for Medical Research, 69120 Heidelberg, Germany.