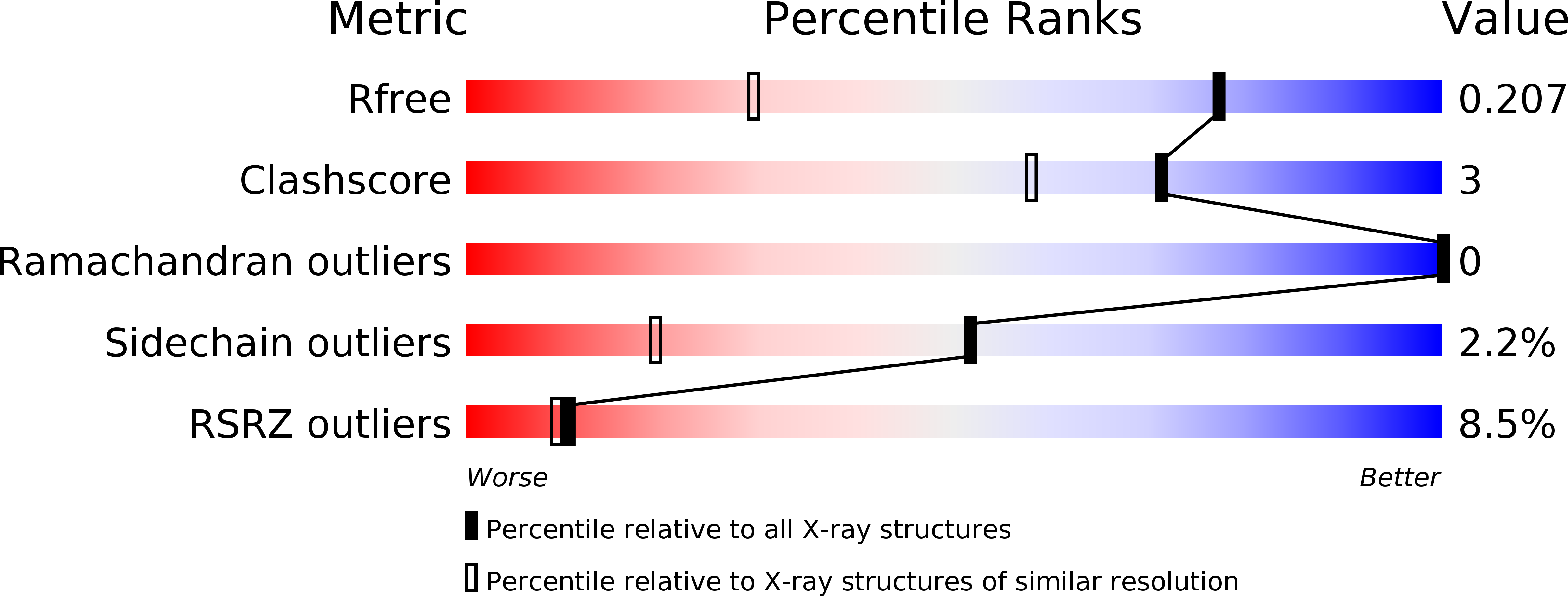
Insight to the residue in P2 position prevents the peptide inhibitor from being hydrolyzed by serine proteases.
Xue, G., Xie, X., Zhou, Y., Yuan, C., Huang, M., Jiang, L.(2020) Biosci Biotechnol Biochem 84: 1153-1159
- PubMed: 32019421
- DOI: https://doi.org/10.1080/09168451.2020.1723405
- Primary Citation of Related Structures:
6XVD - PubMed Abstract:
Peptidic inhibitors of proteases are attracting increasing interest not only as drug candidates but also for studying the function and regulation mechanisms of these enzymes. Previously, we screened out a cyclic peptide inhibitor of human uPA [Formula: see text] and found that Ala substitution of P2 residue turns upain-1 to a substrate. To further investigate the effect of P2 residue on the peptide behavior transformation, we constructed upain-1-W3F, which has Phe replacement in the P2 position. We determined K D and K i of upain-1-W3F and found that upain-1-W3F might still exist as an inhibitor. Furthermore, the high-resolution crystal structure of upain-1-W3F·uPA reveals that upain-1-W3F indeed stays as an intact inhibitor bind to uPA. We thus propose that the P2 residue plays a nonnegligible role in the conversion of upain-1 to a substrate. These results also proposed a strategy to optimize the pharmacological properties of peptide-based drug candidates by hydrophobicity and steric hindrance. Abbreviations : uPA: urokinase-type plasminogen activator; SPD: serine protease domain; S1 pocket: specific substrate-binding pocket.
Organizational Affiliation:
College of Chemistry, Fuzhou University, Fuzhou, China.